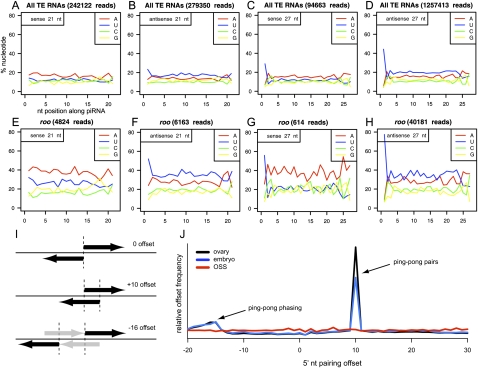
Figure 4.
Sequence properties of TE-siRNAs and TE-piRNAs in OSS cells. (A–D) Aggregate nucleotide composition of all TE-derived reads of length 21 nt (siRNA) or 27 nt (as a representative piRNA size); in all graphs, the x-axis represents nucleotide position along the piRNA, while the y-axis represents the percent nucleotide composition and each position. 5′ U bias is observed for bulk AS-TE-siRNAs as well as S- and AS-TE-piRNAs, but is lacking in bulk S-TE-siRNAs. Note also that S-TE-piRNAs lack any bias for adenine at position 10 (above the adenine bias of neighboring nucleotide positions), as would be expected for ping-pong pairs. Analysis of other TE read sizes are available in Supplemental Figure S3. (E–H) Specific analysis of roo reads shows similar trends. Analysis of all other individual TEs are available in Supplemental Figure S4. (I) Overlap analysis of piRNAs with three hypothetical configurations of distance between 5′ ends of a piRNA and its nearest neighbor on the opposite strand; +10 offset is typical of piRNA ping-pong, while −16 offset is consistent with a phased arrangement of ping-pong pairs (gray arrows depict “missing” piRNAs). (J) Analysis of ovary (black) and 0–2-h embryo (blue) piRNAs reveals strong ping-pong (+10 offset pairs) and modest evidence for a phased ping-pong pairs. No ping-pong is observed for OSS piRNAs (red).