Abstract
Naturally existing biological systems, from the simplest unicellular diatom to the most sophisticated organ such as human brain, are functional self-assembled architectures. Scientists have long been dreaming about building artificial nanostructures that can mimic such elegance in nature. Structural DNA nanotechnology, which uses DNA as blueprint and building material to organize matter with nanometer precision, represents an appealing solution to this challenge. Based on the knowledge of helical DNA structure and Watson-Crick base pairing rules, scientists have constructed a number of DNA nanoarchitectures with a large variety of geometries, topologies and periodicities with considerably high yields. Modified by functional groups, those DNA nanostructures can serve as scaffolds to control the positioning of other molecular species, which opens opportunities to study intermolecular synergies, such as protein-protein interactions, as well as to build artificial multi-component nano-machines. In this review, we summarize the principle of DNA self-assembly, describe the exciting progress of structural DNA nanotechnology in recent years and discuss the current frontier.
The central task of nanotechnology is to control motions and organize matter with nanometer precision. To achieve this, scientists have intensively investigated a large variety of materials including inorganic materials (e.g. carbon nanotubes) (1), organic molecules (2) and biological polymers (e.g. peptides, RNA and DNA) (3–4) as well as different methods that can be sorted into so-called “bottom-up” and “top-down” approaches. Among all of the remarkable achievements made, the success of DNA self-assembly in building programmable nanopatterns attracts broad attention and holds great promise for building novel designer nanoarchitectures (4). DNA is an excellent nano-construction material because of its inherent merits: First, the rigorous Watson-Crick base-pairing makes the hybridization between DNA strands highly predictable. Second, the structure of the B-form DNA double helix is well-understood; its diameter and helical repeat have been determined to be ~2 nm and ~3.4 nm (i.e. ~10.5 bases), respectively, which facilitates the modeling of even the most complicated DNA nanostructures. Third, DNA possesses combined structural stiffness and flexibility. The rigid DNA double helixes can be linked by relatively flexible single-stranded DNA (ssDNA) to build stable motifs with desired geometry. Fourth, modern organic chemistry and molecular biology have created a rich toolbox to readily synthesize, modify and replicate DNA molecules. Finally, DNA is a biocompatible material, making it suitable for the construction of multi-component nanostructures made from hetero-biomaterials.
The simplest example of DNA self-assembly can be found in almost all living forms in nature: two complementary ssDNA molecules spontaneously hybridize together and form a double stranded DNA (dsDNA) molecule. This process is driven by a number of non-covalent interactions such as hydrogen-bonding, base stacking, electrostatic forces and hydrophobic interactions and strictly obeys the Watson-Crick base-paring rules. Artificially designed DNA self-assembly is utilized by molecular biologists in gene cloning. In this case, the genomic DNA of interest is extended at both ends to have ssDNA overhangs (sticky-ends) complementary to the sticky-ends generated by restriction enzymes on the vector; the genomic DNA can then merge into the digested vector and the nick can be sealed by DNA ligase. However, if more complicated nanostructures are desired, one must employ branched DNA motifs to extend the DNA self-assembly into the second, or even the third, dimension. Such motifs have also been found in nature: the DNA replication fork represents a three-way junction motif and the Holliday (4-arm) junction motif exists as the central intermediate of DNA recombination. In 1982, Nadrian Seeman first envisioned the possibility of combining branched DNA molecules bearing complementary sticky-ends to construct two-dimensional (2D) arrays (5). This proposal, which was later experimentally realized, is considered to be the very first stepping stone on the path leading to structural DNA nanotechnology.
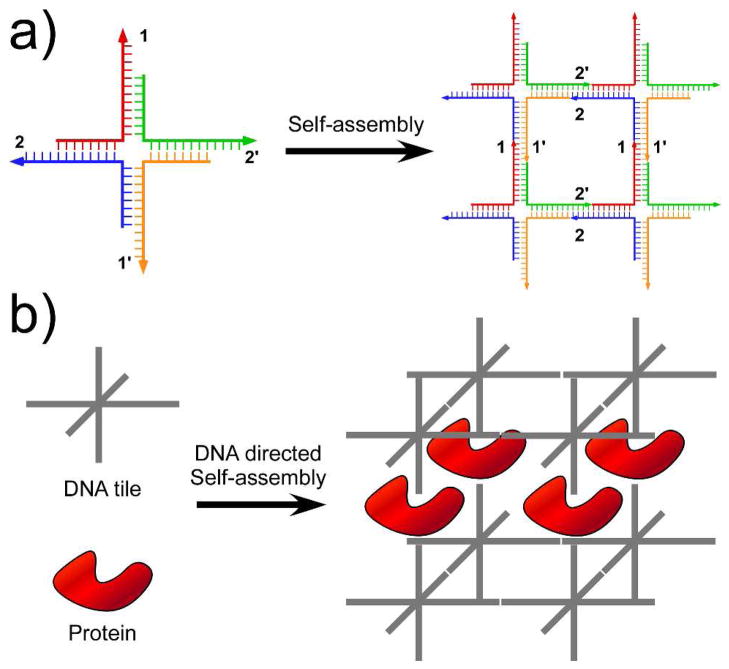
Figure 1a illustrates Seeman’s initial proposal of DNA self-assembly. Here, the basic building block (also known as “tile”) for the DNA 2D array is a 4-arm-junction complex formed by four ssDNA molecules. In addition to the main body, sticky-ends are placed at the termini of each tile with the designated base paring strategy (1 is complementary to 1′ and 2 is complementary to 2′). As a result, the tiles can infinitely grow into a periodic 2D array through sticky-end association. It is intuitive to imagine expanding such self-assembly to the third dimension using 3-, 4-, 5-, or 6-arm junction tiles (Figure 1b, grey tiles). This simple model also illustrates that self-assembly is a hierarchical process: the individual tile formations happen first, and the tile-tile associations follow. Consequently, Seeman proposed the potential application of self-assembled DNA nanostructures as scaffolds to regulate the positioning of other macromolecules in 3D. As shown in Figure 1b, the protein molecules can be assembled parallel to each other with well-defined spatial spacing, directed by the formation of periodic 3D DNA lattice. Once realized, these lattices could provide a universal means to crystallize macromolecules and facilitate subsequent structural determination using X-ray diffraction.
Figure 1.
Principle and application of DNA self-assembly, as proposed by Ned Seeman. (a) Principle of DNA self-assembly: combining branched DNA nanostructures with sticky-ends to form 2D arrays. Arabic numbers indicate base-paring strategies between sticky-ends (1 is complementary to 1′, etc.) (b) Protein crystallization templated by DNA 3D self-assembly.
1. DNA tiles and periodic DNA arrays
The past decade witnessed the fast evolution of structural DNA nanotechnology (4, 6–10). A large variety of DNA tiles with different geometries and topologies has been constructed with excellent yield. Generally speaking, the creation of a novel DNA motif usually requires the following steps: 1) Structural modeling: physical and/or graphic models are used to help the design of a new DNA motif. These models are based on the basic knowledge of DNA helical structures (e.g. helical pitch, diameter, base stacking, phosphodiester bond length, etc.) and are built to give the designer straightforward information (e.g. size, shape and stability) about the designed architecture. The most important consideration for the structural design is to minimize the free energy of the final DNA complex to promote spontaneous assembly. In other words, all the DNA strands involved should stay “comfortably” in the final structure (i.e. no over-stretched bonds, no over-bent helixes, etc.). Today, computer programs (e.g. Mfold, GIDEON, Tiamat and Nanoengineer-1) are designed to aid modeling and free energy predictions (11–14). A complicated structure is deemed “good” or “bad” based on empirical parameters; though even the most experienced designer cannot be assured of successful assembly at this stage. Fortunately, with a large and growing number of DNA nanostructures available, we’re now richly equipped to design new structures by adapting, jointing or scissoring known structures. 2) Sequence design: in this step, specific sequences are assigned to all ssDNA molecules in the model. A general rule of sequence design is to minimize sequence symmetry in the branched structure to avoid possible undesired pairing between participating strands and mobility of the junction points. It is important to point out that sequence symmetry is only avoided within each individual strand but can be allowed between different strands at the symmetric positions in the same tile. The sequence designing process is now automated by computer programs such as SEQUIN (15), Tiamat (13) and Uniquimer (16). It is worth to mention that these programs are designed to break sequence symmetry throughout the whole DNA construct. When symmetry is desired within the system, the sequence for each DNA strand should be designed separately. 3) Experimental synthesis of the DNA nanostructure: Briefly, the oligonucleotides with designated sequences are synthesized by a DNA synthesizer, purified via electrophoresis or chromatography, mixed together at the stoichiometric molar ratio in a near-neutral buffer containing divalent cations (usually Mg2+), heated to denature and then gradually cooled to allow the ssDNA molecules to find their correct partners and adopt the most energy-favorable conformation (i.e. self-assembly). 4) Characterization of the DNA nanostructure: A number of assays can be used to test whether the desired nanostructure forms as designed. The most commonly used methods include non-denaturing gel electrophoresis (tests the integrity of individual tiles), Ferguson study (suggests the shape and size of the tiles) (17), hydroxyl radical autofootprinting (determines the junction point) (18), atomic force microscopy and electron microscopy (AFM and EM, directly visualizes the formation of DNA lattices or large 3D constructions).
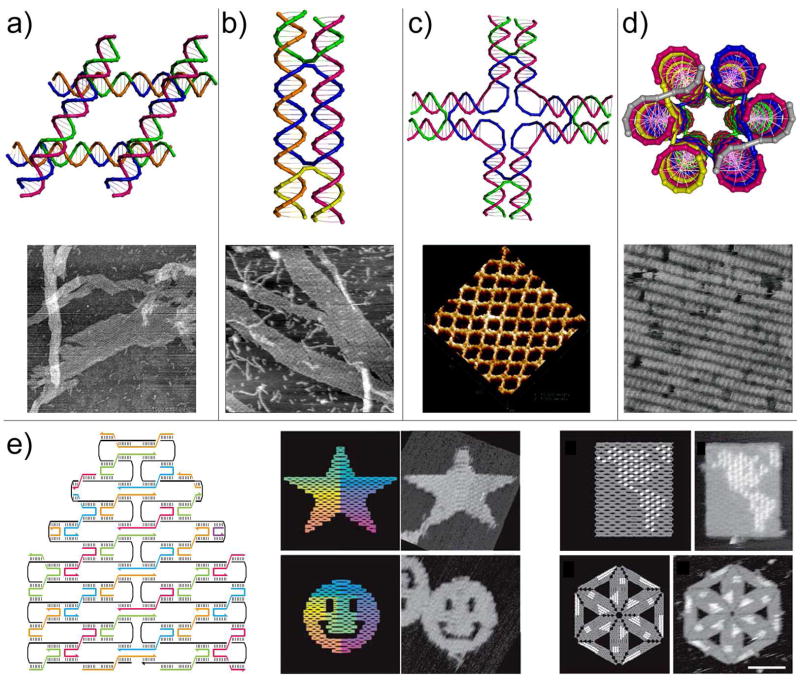
Representative DNA tiles and corresponding periodic self-assembly results are summarized in Figure 2. Closely related to Seeman’s initial proposal, Mao et al constructed parallelogram DNA junctions (Figure 2a) by covalently joining four Holliday junctions together (19). These tiles can self-assemble into 2D arrays with rhombus-shaped cavities. Applying the concept of “tensegrity” and taking advantage of the natural conformation of the Holliday junction, Mao’s group constructed a triangle tile composed of three Holliday junctions (20). Seeman and colleagues first created a series of DNA double crossover (DX) motifs by joining two parallel helices together through strand exchange (21). These DX molecules were later modified to carry proper sticky-ends and successfully self-assembled into DNA 2D arrays without observable cavities (22). Using a similar principle, multi-crossover molecules, including triple crossover (23) and four, eight and twelve helix planar tiles (24, 25), were synthesized to self-assemble into either DNA nanotubes or 2D lattices. Yan and Labean reported the construction of a cross-shaped motif, or so-called 4×4 tile (four 4-arm-junctions linked together through flexible dT4 linkers, Figure 2c), that was used to template the formation of conductive nanowires or protein 2D arrays (26). Mao’s group took advantage of the C4 symmetry of the 4×4 tile and introduced sequence symmetry to the DNA strands in the same tile. By doing this, they effectively minimized the number of unique DNA strands required and reduced possible experimental errors (27). They later applied the same principle to the design of other symmetric tiles such as three-point and six-point star motifs (28, 29). Nicely formed 2D lattices spanning up to square-millimeters were observed in these works, which can be attributed to reduced distortion conferred by the sequence symmetry. In addition to the above-mentioned planer tiles, three-dimensional six-helix (Figure 2d) and three-helix DNA bundle tiles were built (30, 31). These tiles can form nanotubes with fixed diameters, periodic 2D lattices and potentially 3D crystals.
Figure 2.
Models of some representative DNA tiles and their assemblies into periodic 2D arrays. (a) Parallelogram DNA tile formed by joining four Holliday junctions in parallel. (b) Double helix (DX) tile formed through strand exchange between two DNA duplexes. (c) A cross-shaped tile with four arms (4×4 tile); each arm represents a four-arm junction. (d) Six-helix bundle tube tile viewed from the end of the tube. (a)–(d) Representative AFM images of the 2D arrays were shown below the corresponding cartoon models. (e) DNA origami. Left: principle of DNA origami: folding long ssDNA into shapes by multiple helper strands. Middle: Star and smiley face DNA origami tiles self-assembled by folding a 7-kb ssDNA with over 200 helper strands. Right: Hairpin loops (white dots) can be introduced to certain helper strands to accurately display designated geometries on the fully addressable origami tiles.
An exciting breakthrough was made by Rothemund as he first presented “scaffolded DNA origami” (Figure 2e) which was formed by folding M13mp18 genomic DNA (7 kilobase) into desired shapes (~100 nm in diameter) with the help of over 200 short DNA strands (known as helper strands) (32). The versatility of the system was demonstrated by the formation of five arbitrary geometries including rectangles, squares, triangles, stars and smiley faces. DNA origami is a fully addressable molecular pegboard with over 200 six-nanometer pixels because each helper strand at a specific position has a unique sequence. The addressability was demonstrated by introducing DNA hairpins to the helper strands at certain positions to display arbitrary characters (e.g., “DNA”) or shapes (e.g., diagram of DNA double helix) on the origami tile. Individual origami tiles can be further assembled into 2D arrays through base-pairing between extended helper strands on one tile and the unpaired scaffold DNA (7-kb ssDNA) on another. “Scaffolded origami” self-assembly is a powerful tool to generate finite addressable nanostructures in both 2D and 3D. For example, Shih and colleagues constructed DNA octahedrons (33) and nanotubes (34) with fixed dimensions using either artificial or natural ssDNA as scaffolding strand. More complicated patterns can be potentially accessed by selectively combining homogeneous or heterogeneous origami tiles through programmed connectivity, as will be discussed in the next section.
2. Programmable connectivity between DNA tiles
So far we have reviewed a number of DNA tiles with various shapes and sizes. As we mentioned in the introduction of this review, sticky-ends are just as important as the main body of each DNA tile in controlling the final products of self-assembly. While the geometry of the tile defines the repeating unit of the nanopattern, the sticky-end pairing strategy defines the connectivity between tiles; directly determining the size, periodicity and addressability of the nanopattern. It has been shown that one can even change the morphology of the nanopattern from planar array to curled-up nanotube by altering the sticky-end design of each tile (25, 26). Therefore, designing programmable connectivity between DNA tiles allows us to create more complicated nanopatterns than the previously-discussed infinite periodic arrays.
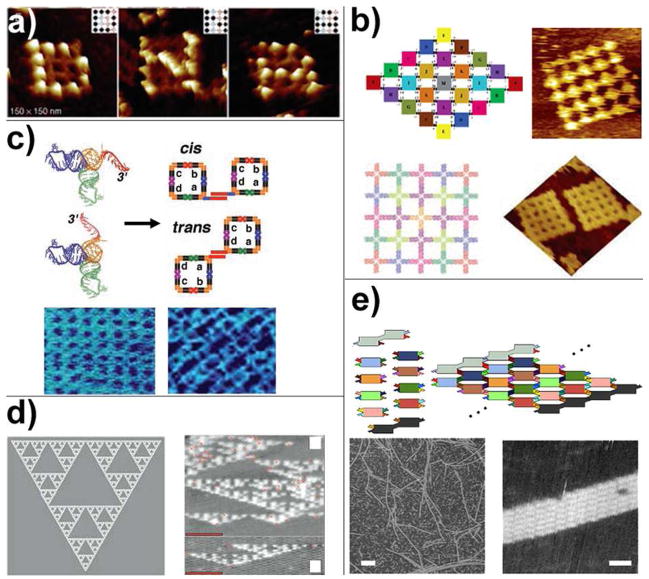
Two independent studies by Yan’s and LaBean’s groups reported the construction of finite-sized arrays made of ten and sixteen 4×4 tiles (Figure 3a), respectively (35, 36). Unlike the assembly of infinite periodic arrays, each tile used in synthesizing finite arrays has unique sticky ends to define its specific location in the final product. As a result, the finite-sized array generated by this method is fully addressable. Both groups demonstrated this by attaching streptavidin proteins on certain tiles. As shown in Figure 3a, the streptavidin molecules were assembled on the sixteen-tile lattices to display letters “D”, “N” and “A”. Such stepwise assembly, although featuring considerably high yield (70%–80%), is not ideal to construct larger finite-sized arrays because the increasing array size requires extra sets of unique sequences, making the system more expensive and error-prone. To address this problem, Yan and coworkers used tiles with symmetric sticky-ends to build the geometrically symmetric finite-sized arrays (Figure 3b) (37). Theoretically, to construct a fixed-size 2D array consisting of N tiles with m-fold symmetry, the number of unique tiles required is N/m, if N/m is an integral number, or Int(N/m)+1 if N/m is a nonintegral number. For example, to assemble a two-fold symmetric 5×5 eight-helix tile array, only 13 unique tiles are necessary instead of 25 tiles. Similarly, 7 unique tiles are enough to construct a 25-tile square with C4 symmetry; 4 unique tiles can assemble into a 24-tile hexagon with C6 symmetry. This method effectively simplified the design and experimental procedure to construct complex nanopatterns at the cost of the addressability of such patterns. Another approach to reduce the unique DNA sequences required for the construction of finite-sized arrays was demonstrated by Park et al (36). In their work, the final product was assembled in multiple steps to “reuse” the same sticky-ends on multiple tiles. The multi-step assembly kept the tiles with exposed homogeneous sticky-ends from being assembled at the same time. The undesired assembly was therefore reduced, because the homogenous sticky-ends were already buried inside the assembly intermediates when they met and were less likely to interfere with the correct self-assembly in the next step. However, as is common in multistep synthesis, this procedure is associated with less efficiency (yield ~30%), which may be improved by purification after each assembly step.
Figure 3.
Programmable connectivity between DNA tiles. (a) a sixteen-tile finite-sized DNA array made of cross-shaped tiles. Streptavidins were attached to certain tiles to display letters “D”, “N” and “A” on the arrays. (b) Taking advantage of the geometric symmetry, finite-sized DNA arrays can be assembled from minimal number of unique tiles. Shown here are a two-fold and a four-fold symmetric 25-tile arrays constructed using 13 and 7 unique tiles, respectively. (c) Selective combinations of tecto-RNAs can self-assemble into tecto-squares with different sticky-tail conformations, which further self-assemble into finite and infinite arrays with various cavity and periodicity. (d) Algorithm self-assembly of Sierpinski triangle DNA sheet. (e) Nucleated assembly of fixed-width DNA ribbon.
Jaeger and colleagues described artificial RNA self-assembly using tecto-RNA (a T-shaped RNA motif derived from small RNA motifs present in the ribosomal structures) as the basic building block (Figure 3c) (38). Four non-identical tecto-RNA molecules were held together through kissing loop interactions to form so-called tecto-squares and served as the assembly module in the next step. The 3′ end of each tecto-RNA molecule can be extended to form “sticky-tails” and allow designated association between the tecto-squares. Furthermore, the direction of the sticky-tails can be purposely altered to switch the conformation of the associated double-squares. A diverse expanse of infinite and finite RNA arrays with various shapes, cavities and periodicities were produced. Besides pioneering non-DNA nucleic acid assembly, this work has also provided an excellent example of modularly-designed two-step hierarchical self-assembly.
Algorithm self-assembly is another way to program the connectivity between DNA tiles. The rationale behind this strategy is to use the DNA tiles as basic computing units (e.g., 1 and 0) and program the logic operation command (e.g., XOR) into the sticky-ends. As a result, the self-assembly process is governed by the programmed algorithm and automatically yields a pattern reflecting the computing result, which can be read out by analytical methods such as AFM. Alternatively, algorithm self-assembly can be regarded as generalized and programmed DNA 2D crystallization. It has demonstrated its power in generating some of the most complicated non-periodic DNA arrays to date, such as the Sierpinski triangles constructed by Rothemund et al (Figure 3d), which involved four sets of DX tiles as computing elements to carry out the XOR operation (39). A DX sheet carrying out the AND operation was used by Winfree and coworkers to perform binary counting up to 8 (40). Because early examples of algorithm self-assembly usually suffered from high error rates and low yield, a number of efforts have been made in recent years to improve the assembly efficiency (41, 42). For example, Chen et al introduced additional “proofreading” tiles to reduce the chance of incorrect self-assembly (41). It is worth mentioning that algorithm self-assemblies commonly require nucleation to avoid random, untemplated crystallization. For example, a string of tiles generated by assembly PCR was employed as “boundary tiles” to define the initial input for the assembly of the Sierpinski triangle sheet (39). Origami tile boundaries are excellent candidates for such nucleation purposes because of their characteristic fixed lengths and full addressability (42). Such nucleated self-assembly has drawn lots of attention itself. The most well-known example of this is perhaps DNA origami, which uses viral genomic DNA as the nucleation strand (32). To further understand the kinetics and have better control of nucleated DNA self-assembly, Schulman and Winfree constructed a series of “zigzag ribbons” (Figure 3e) with fixed width using two sets of dimeric DX tiles as seed tiles to provide crystallization nuclei and to remove the nucleation energy barrier (43).
3. Construction of 3D nanostructures
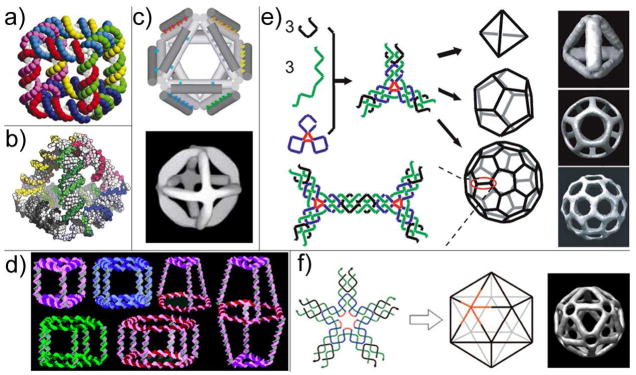
With the inevitable success of structural DNA nanotechnology, scientists now are able to construct almost any 2D patterns we can imagine using DNA self-assembly. However, we are still far away from the goal of using DNA as scaffolding to enable structure determination of proteins and mimicking the elegance of naturally-occurring self-assembly systems. To achieve (or perhaps first get closer to) those objectives, we must develop efficient and reliable assembly strategies to construct 3D nanostructures. Conceivably, there should be no conceptual difference between 2D and 3D self-assembly, which means the principles behind 2D self-assembly can still apply in 3D. Seeman and his fellow researchers first experimentally constructed DNA objects that are topologically equivalent to a cube (Figure 4a) and a truncated octahedron (44, 45). In these works, the DNA polyhedrons are comprised of closed, interlocked DNA rings; each edge of the polyhedron is made of duplexed DNA and each vertex represents an immobile multi-arm junction. These polyhedrons were synthesized in a step-wise manner through a series of ligation and purification steps to ensure the yield of the correct assembly products. Though labor-intensive and inefficient, these early successes proved the capability of DNA to serve as the scaffold for 3D construction.
Figure 4.
DNA self-assembled 3D nanoarchitectures. (a) Model of a DNA cube. (b) Model of a DNA tetrahedron. (c) Model (top) and cryo-EM image (bottom) of a DNA octahedron. (d) A library of 3D prisms and cubes are assembled from cyclic and single-stranded DNA molecules with organic vertices. (e) DNA tetrahedron, dodecahedron and buckyball each self-assembled from a single symmetric three-point star tile. (f) DNA icosahedron self-assembled from a five-point star tile.
A burst of new 3D DNA motifs have emerged over the past five years. Through elegantly simple design and experimentation, Turberfield’s group constructed a series of DNA tetrahedrons (Figure 4b) with different dimensions using as few as four DNA strands (46, 47). These structures were assembled exclusively through hybridization of participating ssDNA molecules (i.e., no ligation is necessary) and featured high yield (>95% at 50 nM DNA concentration). In a latter study, hairpin loops were incorporated on the edges to build tetrahedrons with variable dimensions (48). The hairpin loops can be opened by additional “fuel strands” and re-closed by “anti-fuel strands” based on the strand displacement mechanism. Together with the success of encapsulating a cytochrome c protein inside a tetrahedron cage (detail in the next section) (49), the work of this same group has established solid steps toward controlled drug release using DNA cages as delivery vehicles. Shih, Quispe and Joyce built a DNA octahedron by folding a 1.7-kb ssDNA with 5 short DNA helper strands (Figure 4c) (33). The edges of this octahedron incorporate DX or PX (paranemic crossovers) motifs (50), which are nearly twice as rigid as single-duplex DNA. Moreover, the main component of the tetrahedron (1.7-kb ssDNA) is clonable, making it one of the first examples of replicable DNA nanostructures (see discussion in section 6). The assembly products were examined by cryo-EM, which clearly revealed the successful folding of the DNA strands into an octahedron motif.
Sleiman introduced a novel stepwise assembly method to construct 3D DNA prisms (Figure 4d) (51). In the first step, single-stranded and cyclic DNA molecules were generated by solid-phase DNA synthesis and subsequent DNA-templated chemical ligation. The geometry of these cyclic ssDNA molecules was defined during DNA synthesis by controlling the length of the oligonucleotide and the number of “vertex organic molecules” coupled. As a result, a library of DNA polygon structures (e.g., triangle, rectangular, pentagon, and hexagon) with single-stranded DNA sides and “vertex organic molecules” at the corners was generated. In the next step, two of those DNA polygons were assembled together to serve as the top and bottom faces of a 3D DNA prism by linking strands and the construction was finalized by the addition of rigidifying strands to strengthen the vertical edges. This modular design and combinatorial assembly strategy enabled the fabrication of a large number of 3D DNA cages including DNA cube, homo-, hetero- and bi-prisms. Dynamic DNA cages were realized by the addition and displacement of rigidifying strands with different lengths.
Recently Mao’s group reported the hierarchical assembly of symmetric DNA polyhedrons including tetrahedra, dodecahedra and buckyballs (Figure 4e) (52). The basic building blocks of these polyhedrons are symmetric three-point-star tiles with sticky-ends. Two factors were varied in order to selectively assemble a certain kind of polyhedron with optimal yield: first, the length of the poly-dT linkers inside the three-point-star tile was varied to control the flexibility of the tile; and second, the 3D nanostructures were assembled at different concentrations to control the number of tiles inside one polyhedron. The same group later assembled an icosahedron using a five-point-star tile by the same principle (Figure 4f) (53).
4. DNA tile-directed assembly of multi-component nanoarchitectures
Self-assembled designer DNA nanoarchitectures hold great promise as scaffolds to organize other nano-scale entities. Such DNA-directed assembly strategies gave rise to multi-component nanoarchitectures in which macromolecules are displayed on the DNA lattices with well-controlled intermolecular distances. This opens up exciting opportunities for both fundamental studies of distance-dependent molecular interactions and practical applications like biosensing, drug delivery, DNA-templated chemistry and crystallization. The success of constructing such hetero-material nanostructures has already generated a considerably large impact in materials science, chemistry, physics and biology; making structural DNA nanotechnology a cutting-edge interdisciplinary field.
There are generally two ways to “functionalize” DNA nanostructures by other molecular species. The functionality could come from: 1) covalently attached functional groups or molecules (e.g., thiol, amino and carboxylic groups or biotin) that can chemically link to their specific targeted molecular species, such as gold nanoparticles and proteins; and 2) extensions of single-stranded or stem-loop DNA or RNA probes that can hybridize and capture targets with complementary sequences or through specific aptamer-target binding. Both methods have been intensively applied in DNA-directed self-assembly.
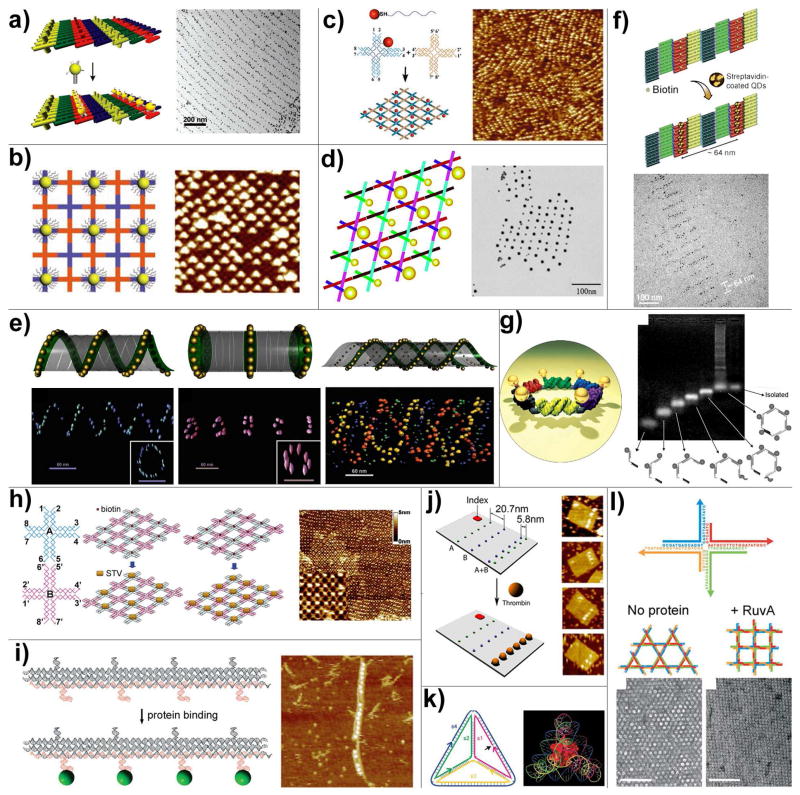
Among all materials that can be organized on DNA scaffolds, inorganic nanomaterials (e.g., carbon nanotubes, metallic and semiconducting nanoparticles) and biomolecules (e.g., nucleic acids, proteins and antibodies) are the most thoroughly investigated substances because of their interesting physical and biological properties and functions. Through DNA hybridization, gold nanoparticles (AuNP) modified by multiple copies of ssDNA were organized on the surface of periodic DNA nanoarrays (self-assembled from DX tiles or cross-shape tiles) bearing complementary probes (Figure 5a and 5b) (54, 55). In these approaches, the DNA arrays were assembled first and deposited on the surface, followed by the addition of ssDNA coated AuNPs. Using purified 1:1 conjugate of AuNP and 100-mer ssDNA, the AuNP arrays can be obtained in a one-pot annealing process (Figure 5c), as demonstrated by Yan’s group (56). Short poly-dT strands were coated on the AuNPs to stabilize them in the high salt buffer environment that is required for DNA self-assembly. Such DNA mono-functionalized AuNPs were later proved to be highly useful to generate more complicated nanoparticle patterns. For example, Seeman and colleagues constructed checkerboard-like 2D nanoparticle arrays by incorporating 5- and 10-nm AuNPs modified each by a single DNA strand into robust triangle-shaped DNA motifs (Figure 5d) (57). Recently, Yan and Liu demonstrated that by the attachment of ssDNA to gold nanoparticles, nanotubes of various 3D architectures can form, ranging in shape from stacked rings to single spirals, double spirals, and nested spirals (Figure 5e) (58). In this case, the nanoparticles are active elements that control the preference for specific tube conformations through size-dependent steric repulsion effects. Templated by biotinylated DNA 2D lattices or nanotubes, streptavidin-modified quantum dots were turned into highly ordered arrays through biotin-streptavidin interaction (Figure 5f) (59, 60). Patterning a discrete number of nanoparticles in a deliberately-designed, finite architecture is critical in nanocircuit fabrication. An endeavor undertaken by Sleiman et al, shown in Figure 5g, resulted in a hexagonal pattern of AuNPs templated by DNA designed using sequential self-assembly methodology (61). Six AuNP-conjugated DNA building blocks were synthesized and assembled in a sequential manner to achieve a hexagon pattern of AuNP. To achieve more reliable AuNP patterning on DNA nanoscaffolding, a stronger linkage between AuNPs and DNA strands is desired. Liu and co-workers reported a new strategy to prepare AuNPs monofunctionalized with lipoic acid-modified DNA oligonucleotides (62). These conjugates were further selectively mixed with other DNA strands and assembled into fixed-sized DNA nanostructures (rectangular origami tiles) carrying a discrete number of AuNPs at desired positions.
Figure 5.
DNA directed assembly of multi-component nanoarrays. (a) Organization of 5 nm AuNPs on DNA DX lattices. (b) Periodic 5 nm AuNP nanoarrays with well controlled interparticle distances templated by 2D DNA nanogrids. (c) DNA mono-modified 5 nm AuNPs directly participate in the self-assembly process and yield periodic nanoparticle arrays. (d) 2D periodic array of 5- and 10-nm AuNPs generated by incorporating DNA mono-modified AuNPs into robust triangle-shaped DNA motifs. (e) Controlled self-assembly of DNA tubules through integration of AuNPs. The assembly results in 3D nanoparticle architectures such as single-spiral tube (left), stacking ring tube (middle) and interlocking double-spiral tube (right). The schematic views are placed above corresponding electron tomographic images. (f) Quantum dots organized on DNA DX lattices through biotin-streptavidin interaction. (g) Discrete hexagonal AuNP array displayed on a DNA hexagon consisting of six non-identical molecules each with two ssDNA arms linked by an organic molecule. (h) Programmable streptavidin 2D arrays formed on biotinylated DNA lattices. (i) 1D thrombin array assembled by incorporating anti-thrombin aptamers into linear TX DNA array. (j) Selective binding of thrombin proteins to the bivalent anti-thrombin aptamers displayed on the surface of rectangular origami arrays. (k) A cytochrome c protein trapped inside a DNA tetrahedron cage. (l) The binding of RuvA to Holliday junction tiles alters the assembly product from Kagome-type lattice (left) to square-planar lattice (right).
Figure 5h–5l shows representative examples of protein nanopatterns generated through DNA-directed assembly. A two-tile system was used to build programmable streptavidin arrays (Figure 5h) by Park et al (63). The density of the protein on the array was controlled by assembling the DNA lattice with half or all tiles biotinylated. Liu and coworkers first demonstrated the use of aptamers, which are DNA or RNA oligonucleotides with specific molecular binding affinities, to direct the assembly of thrombin onto sites on the linear three-helix tile arrays (64). As illustrated in Figure 5i, anti-thrombin aptamers were incorporated into three-helix tiles that could self-assemble into 1D tracks. This is a modular design with high adaptability. For example, by using aptamers with distinct specificities at different positions of an addressable DNA scaffold, addressable protein arrays can be constructed, as achieved by Chhabra et al (65). By coupling the molecular binding events with detectable signal outputs, both fluorescently-labeled and label-free DNA-nanoarray-based multiplexed biosensors were created (66, 67). Inspired by the “multivalent binding” phenomenon in biology, Yan’s group recently used DNA scaffolds as tunable platforms to generate bivalent anti-thrombin aptamers with stronger binding affinity (68). Figure 5j shows the selective binding of thrombin molecules to the bivalent aptamer lines with optimal interline distance on a rectangular origami tile. Chemical modification is another way to incorporate protein into DNA nanostructure. As demonstrated by Turberfield’s group, a cytochrome c protein was covalently linked to a ssDNA and the protein-DNA conjugate was added to the self-assembly mixture in place of the corresponding naked DNA strand, resulting in a protein molecule encapsulated inside the DNA tetrahedron (Figure 5k) (49). Using the same concept and DNA cage, but site-specific “click” chemistry to link protein and DNA, Distefano’s group constructed a nanostructure consisting of four green fluorescent proteins and one DNA tetrahedron (69). Turberfield and co-workers demonstrated the binding of RuvA, a Holliday junction binding protein to a 2D Holliday junction lattice (70). As illustrated in Figure 5l, when RuvA bound to the building blocks during the self-assembly process, the lattice showed a square-planar configuration rather than the original kagome lattice. This shows that not only can DNA be used to create ordered protein arrays, the protein molecules can also play an active, decisive role dictating the shape of the DNA tile lattices.
5. Replicable DNA nanostructures
Massive parallel construction is one of the key features of the bottom-up fabrication of nano-materials. However, it is always true that the overall yield of self-assembled products depends on the amount of starting material. The challenge, therefore, arises when large quantities of such nano-materials are demanded. Nature seems to have a perfect solution to this dilemma. All living cells, which are delicate self-assembled systems, have the impressive ability to self-replicate to allow the sustainability of the species. Such replication features extraordinary fidelity to ensure the correct inheritance of biological information. This is achieved by a cooperative and sophisticated enzyme network that is dedicated to DNA replication, repair and recombination. Extensive research has been conducted to understand self-replication phenomena in nature; but can we mimic this elegant biological process to benefit nanoscience and nanotechnology by replicating self-assembled artificial nanostructures?
The inherent property of DNA as a replicable molecule renders DNA nanostructures an extra layer of charm. The idea of replicating DNA nanostructures was initially proposed by Seeman in the early 90’s (71). Von Kiedrowski replicated a three-point-star motif by chemical methods, in which the parental nano-motif serves as the template to direct the chemical ligation of non-identical DNA strands to form the next generation of the nano-motif (72). Another work by Shih et al involves a 1.7-kb ssDNA that was used to assemble a DNA octahedron with the help of five short DNA strands (Figure 4c); this molecule was cloned into a bacterial plasmid. A nicking endonuclease was used to digest the cloned double-stranded plasmids after amplification to obtain the 1.7-kb ssDNA (33). This is an inspiring accomplishment, although the long strand did not form a complete nanostructure without the aid of the short helper strands. Using rolling-circle amplification (RCA)-based enzymatic methods, Yan and colleagues recently reported the replication of a DNA Holliday junction and a PX DNA molecule (73, 74). In these cases, ssDNA capable of folding into a designated nanostructure (sense strand) was first ligated to form a circular molecule, which would serve as template for RCA. The rolling circle polymerase (e.g., phi29 polymerase) and DNA primer were then added to generate long ssDNA with tandem repeats of complementary segments of the sense strand (antisense strand). The monomer form of antisense strand was obtained through restriction enzyme digestion. Exact copies of sense strands were generated through one additional repeat of the above process using the circularized antisense strand as template. These works provided proof-of-concept demonstration that artificial DNA nanoarchitectures are replicable materials.
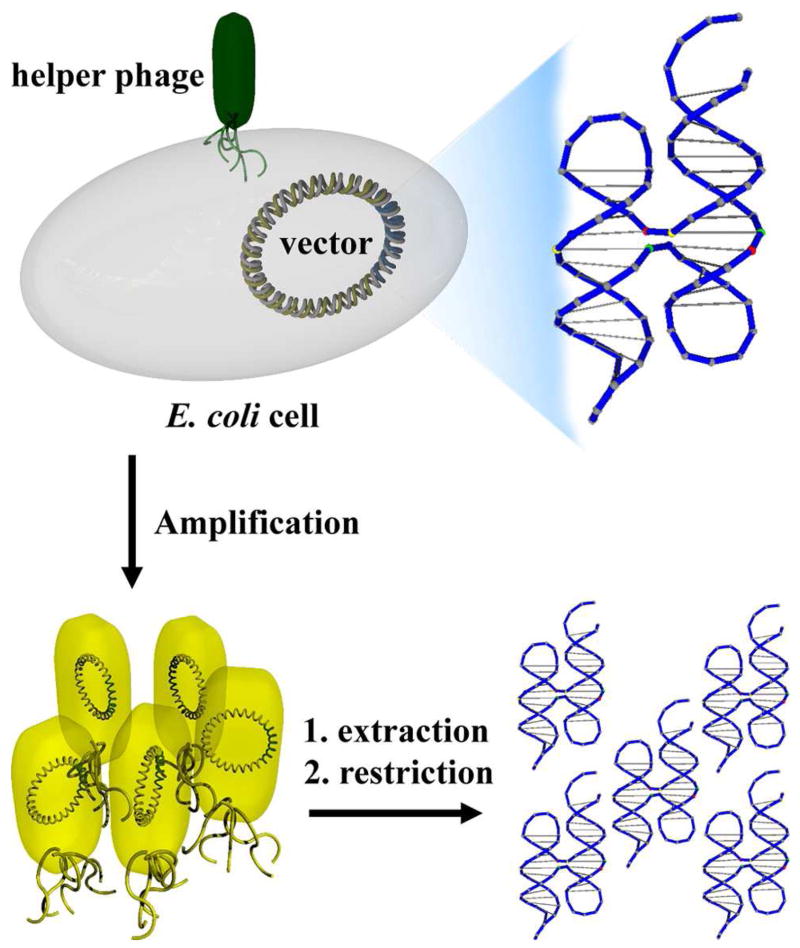
Taking full advantage of naturally-existing DNA replication machinery, research groups led by Yan and Seeman recently reported a system where DNA nanostructures folded from a ssDNA molecule can be readily amplified by bacterial cells and viruses (75). In this work, ssDNA capable of folding into a designated nanostructure was inserted into a double-stranded vector called a phagemid, transformed into Escherichia coli cells and amplified in vivo with the assistance of helper phage (Figure 6). Here the phagemid worked as a vehicle to shuttle the ssDNA nanostructure in and out of the bacteria. When the helper phage was present, the phagemid, in its single-stranded form, was packed in phage particles and secreted into the culture medium. As a result of cell growth, the “nanostructure engineered” phagemid was exponentially amplified, leading to a high copy number of DNA nanostructures after DNA extraction, restriction and re-annealing. Improved replication efficiency was achieved when this strategy was compared to the in vitro replication methods developed by the same group (73, 74). This amplification requires only a small amount of ssDNA nanostructures (sub-picomole) to start with, and can be almost infinitely scaled up, simply by growing larger cultures of cells. Moreover, this research implies that cloned DNA nanostructures can possibly survive within the cellular environment, which suggests that in vivo artificial self-assembly is possible. (See next section for more discussion).
Figure 6.
Schematic illustration of the in vivo replication of DNA Holliday junction structure. The DNA junction was inserted into a phagemid vector, transformed into E. coli cells, and then replicated in the presence of helper phage. The single-stranded phagemid DNA molecules were packed and secreted into the culture medium. After simple post-treatment like DNA extraction and restriction, high copy numbers of cloned nanostructures can be obtained as a result of the exponential replication of phagemid vectors in bacteria cells.
6. Challenges and Outlook
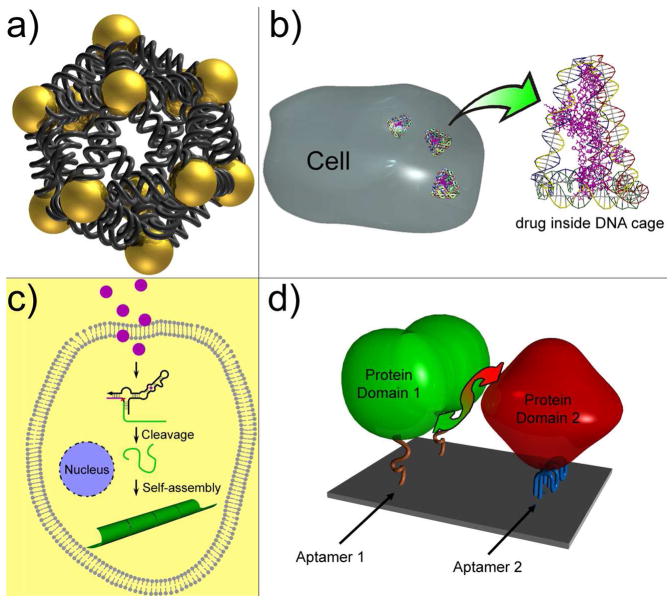
The fast-evolving realm of structural DNA nanotechnology has already shown its power in manufacturing programmable nanoarchitectures with rationally-designed functionality and nanometer precision in addressability. Numerous exciting advances and breakthroughs have given us the precedents and confidence to construct complicated DNA nanostructures with precise control over their geometries, periodicities, chiralities and topologies. However, there are still many challenging opportunities and open questions. First, the mechanism of DNA self-assembly (e.g., what determines the error rates in self-assembly; how to predict the outcome of self-assembly from a given set of DNA strands; can the self-assembly product be deliberately altered via the controlled annealing process; etc.) has not yet been completely elucidated. Better understanding of these topics would not only shed light on the physical chemistry aspects of self-assembly systems, but also would aid future novel design and construction endeavors. Second, our ability to carry out 3D construction of discrete DNA nanostructures seems mature, but 3D-DNA-nanostructure-directed self-assembly has barely been explored. Intuitively, we should be able to use DNA cages to direct the assembly of several short peptides to simulate their native spatial orientations in the protein complex and artificially create a functional enzyme center. Another interesting objective may be to pursue DNA-directed 2D or 3D assembly of discrete nanoparticle patterns (Figure 7a) in order to study their plasmonic interactions. Many expect that unique and advantageous optical and plasmonic properties will emerge from carefully- designed nanostructure configurations and become additional design variables exploitable to yield supplementary functionality. The self-assembly of designed 3D DNA crystals, which is yet to be realized, would enable scaffolded 3D protein crystallization as proposed by Seeman. Third, it is a grand challenge to create more sophisticated functional DNA nanodevices. For example, many DNA-based nano-motors (e.g., tweezers, walkers, gears, etc.) have been constructed and their motions have been confined on DNA tracks (76–78). (These exciting accomplishments have been comprehensively summarized in several review articles. We did not discuss them here in detail for the article-length consideration.) However, none of these devices so far can carry out a mission to load, transport and unload a macromolecular cargo, which is a common feature of the biological motor complexes (e.g., ribosome) in nature. Fourth, the merging of DNA self-assembly, a bottom-up approach together with top-down methods such as dip-pen lithography (79) and molecular combing (80), would bring a new wave of breakthroughs in fabricating DNA-based nanoelectronics, such as nanocircuits. Finally, current DNA self-assembly almost exclusively takes place in vitro. To maximize the outreach of DNA nanotechnology and take full advantage of the biocompatibility of DNA, it would be ideal to construct and deliver functional DNA nanostructures in vivo. Towards this end, the very first consideration would be the structural integrity of the DNA nanostructures in biological environments. Yan and Seeman’s work suggests that simple DNA nanostructures, such as Holliday junctions, can possibly survive in bacterial cells (75). However, cellular tolerance limitations to foreign inserts and the degree of comparability between eukaryotic and bacterial are unknown. Using nanostructures made from modified or unnatural nucleic acids and with nuclease resistance could help improve their chemical stability in vivo (81, 82). Once a reliable in vivo nucleic acid self-assembly method is established, many medical applications, such as in vivo diagnostic and DNA-mediated drug delivery (Figure 7b) could be realized. One day, DNA nanotechnologists may assemble their nano-patterns or operate nano-devices inside living cells to sense a pathogen or kill a cancer cell (Figure 7c). Other biochemical research opportunities may arise as well. It is conceivable that, under proper external selection pressure, novel DNA nanoarchitectures or functional DNA nanodevices may be generated by means of in vivo evolution instead of rational design. These nanodevices could be used to trigger protein-protein interactions (Figure 7d), promote cell growth, or regulate gene expression.
Figure 7.
Perspective applications of DNA nanotechnology. (a) Assembly of discrete 3D nanoparticle structure to study the inter-paticle plasmonic effect. Shown here is a DNA icosahedron with nanoparticle at each vertex. (b) DNA mediated drug delivery. (c) Small molecule triggered in vivo DNA self-assembly for medical applications. For example, the binding between taken up molecule and an allosteric DNAzyme can trigger the cleavage of the substrate DNA. The released DNA segment can then self-assemble into nanotube, which can be visualized as a pathogen marker or directly lead to cell death. (d) DNA nanochip displaying two aptamers can bring two protein subunits into proximity and therefore induce protein-protein interaction.
Acknowledgments
We thank Carole Flores for proof reading the manuscript and Yonggang Ke for the courtesy images used in TOC graphic.
Abbreviations
- ssDNA
single-stranded DNA
- dsDNA
double-stranded DNA
- 1D
one-dimensional
- 2D
two-dimensional
- 3D
three-dimensional
- DX
double crossover
- TX
triple crossover
- PX
paranemic crossovers
- PCR
polymerase chain reaction
- AuNP
gold nanoparticle
- RCA
rolling circle amplification
Footnotes
This work was supported by grants from the National Science Foundation (NSF), the Army Research Office (ARO), and the Technology and Research Initiative Fund from Arizona State University to Y.L. and by grants from NSF, ARO, Air Force Office of Scientific Research, Office of Naval Research, and the National Institute of Health to H.Y.
References
- 1.Odom TW, Huang JL, Kim P, Lieber CM. Structure and electronic properties of carbon nanotubes. J Phys Chem B. 2000;104:2794–2809. [Google Scholar]
- 2.Theobald JA, Oxtoby NS, Phillips MA, Champness NR, Beton PH. Controlling molecular deposition and layer structure with supramolecular surface assemblies. Nature. 2003;424:1029–1031. doi: 10.1038/nature01915. [DOI] [PubMed] [Google Scholar]
- 3.Columbo G, Soto P, Gazit E. Peptide self-assembly at the nanoscale: a challenging target for computational and experimental biology. Trends Biotechnol. 2007;25:211–218. doi: 10.1016/j.tibtech.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Seeman NC. DNA in a material world. Nature. 2003;421:427–431. doi: 10.1038/nature01406. [DOI] [PubMed] [Google Scholar]
- 5.Seeman NC. Nucleic acid junctions and lattices. J Theor Biol. 1982;99:237–247. doi: 10.1016/0022-5193(82)90002-9. [DOI] [PubMed] [Google Scholar]
- 6.Deng ZX, Lee SH, Mao CD. DNA as nanoscale building blocks. J Nanosci Nanotechnol. 2005;5:1954–1963. doi: 10.1166/jnn.2005.504. [DOI] [PubMed] [Google Scholar]
- 7.Turberfield AJ. DNA as an engineering material. Phys World. 2003;16:43–46. [Google Scholar]
- 8.Lin C, Liu Y, Rinker S, Yan H. DNA Tile based self-assembly: building complex nanoarchitectures. ChemphysChem. 2006;7:1641–1647. doi: 10.1002/cphc.200600260. [DOI] [PubMed] [Google Scholar]
- 9.Feldkamp U, Niemeyer CM. Rational fesign of DNA nanoarchitectures. Angew Chem Int Ed. 2006;45:1856–1876. doi: 10.1002/anie.200502358. [DOI] [PubMed] [Google Scholar]
- 10.Aldaye FA, Palmer AL, Sleiman HF. Assembling materials with DNA as the guide. Science. 2008;321:1795–1799. doi: 10.1126/science.1154533. [DOI] [PubMed] [Google Scholar]
- 11.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Birac JJ, Sherman WB, Kopatsh J, Constantinou PE, Seeman NC. GIDEON, A program for design in structural DNA nanotechnology. J Mol Graphics Model. 2006;25:470–480. doi: 10.1016/j.jmgm.2006.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams S, Lund K, Lin C, Wonka P, Lindsay S, Yan H. Tiamat: a three-dimensional editing tool for complex DNA structures. The 14th International Meeting on DNA Computing; Prague, Czech Republic. 2008. [Google Scholar]
- 14.Nanoengineer-1 is a molecular design program developed by Nanorex, Inc (Bloomfield Hills, MI). http://nanoengineer-1.com/content/
- 15.Seeman NC. De novo design of sequences for nucleic acid structure engineering. J Biomol Struct Dynam. 1990;8:573–581. doi: 10.1080/07391102.1990.10507829. [DOI] [PubMed] [Google Scholar]
- 16.Wei B, Wang Z, Mi Y. Uniquimer: software of de novo DNA sequence generation for DNA self-assembly–an introduction and the related applications in DNA self-assembly. J Comput Theor Nanosci. 2007;4:133–141. [Google Scholar]
- 17.Ferguson KA. Starch-gel electrophoresis-application to the classification of pituitary proteins and polypeptides. Metabolism. 1964;13:985–1002. doi: 10.1016/s0026-0495(64)80018-4. [DOI] [PubMed] [Google Scholar]
- 18.Churchill MEA, Tullius TD, Kallenbach NR, Seeman NC. A Holliday recombination intermediate is twofold symmetric. Proc Natl Acad Sci USA. 1988;85:4653–4656. doi: 10.1073/pnas.85.13.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mao C, Sun W, Seeman NC. Designed two-dimensional DNA Holliday junction arrays visualized by atomic force microscopy. J Am Chem Soc. 1999;121:5437–5443. [Google Scholar]
- 20.Liu D, Wang M, Deng Z, Walulu R, Mao C. Tensegrity: Construction of rigid DNA triangles with flexible four-arm DNA junctions. J Am Chem Soc. 2004;126:2324–2325. doi: 10.1021/ja031754r. [DOI] [PubMed] [Google Scholar]
- 21.Fu T-J, Seeman NC. DNA double-crossover molecules. Biochemistry. 1993;32:3211–3220. doi: 10.1021/bi00064a003. [DOI] [PubMed] [Google Scholar]
- 22.Winfree E, Liu F, Wenzler LA, Seeman NC. Design and self-assembly of two-dimensional DNA crystals. Nature. 1998;394:539–544. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]
- 23.LaBean T, Yan H, Kopatsch J, Liu F, Winfree E, Reif JH, Seeman NC. The construction of DNA triple crossover molecules. J Am Chem Soc. 2000;122:1848–1860. [Google Scholar]
- 24.Reishus D, Shaw B, Brun Y, Chelyapov N, Adleman L. Self-assembly of DNA double-double crossover complexes into high-density, doubly connected, planar structures. J Am Chem Soc. 2005;127:17590–17591. doi: 10.1021/ja0557177. [DOI] [PubMed] [Google Scholar]
- 25.Ke Y, Liu Y, Zhang J, Yan H. A study of DNA tube formation mechanisms using 4-, 8-, and 12-helix DNA nanostructures. J Am Chem Soc. 2006;128:4414–4421. doi: 10.1021/ja058145z. [DOI] [PubMed] [Google Scholar]
- 26.Yan H, Park SH, Ginkelstein G, Reif JH, LaBean TH. DNA templated self-assembly of protein arrays and highly conductive nanowires. Science. 2003;301:1882–1884. doi: 10.1126/science.1089389. [DOI] [PubMed] [Google Scholar]
- 27.He Y, Tian Y, Chen Y, Deng ZX, Ribbe AE, Mao CD. Sequence Symmetry as a Tool for Designing DNA Nanostructures. Angew Chem Int Ed. 2005;44:6694–6696. doi: 10.1002/anie.200502193. [DOI] [PubMed] [Google Scholar]
- 28.He Y, Chen Y, Liu H, Ribbe AE, Mao C. Self-assembly of hexagonal DNA two-dimensional (2D) arrays. J Am Chem Soc. 2005;127:12202–12203. doi: 10.1021/ja0541938. [DOI] [PubMed] [Google Scholar]
- 29.He Y, Tian Y, Ribbe AE, Mao C. Highly connected two-dimensional crystals of DNA six-point-stars. J Am Chem Soc. 2006;128:15978–15979. doi: 10.1021/ja0665141. [DOI] [PubMed] [Google Scholar]
- 30.Park SH, Barish R, Li HY, Reif JH, Finkelstein G, Yan H, LaBean TH. Three-helix bundle DNA tiles self-assemble into 2D lattice or 1D templates for silver nanowires. Nano Lett. 2005;5:693–696. doi: 10.1021/nl050108i. [DOI] [PubMed] [Google Scholar]
- 31.Mathieu F, Liao SP, Kopatscht J, Wang T, Mao CD, Seeman NC. Six-helix bundles designed from DNA. Nano Lett. 2005;5:661–665. doi: 10.1021/nl050084f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rothemund PWK. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 33.Shih WM, Quispe JD, Joyce GF. A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron. Nature. 2004;427:618–621. doi: 10.1038/nature02307. [DOI] [PubMed] [Google Scholar]
- 34.Douglas SM, Chou JJ, Shih WM. DNA-nanotube-induced alignment of membrane proteins for NMR structure determination. Proc Natl Acad Sci USA. 2007;104:6644–6648. doi: 10.1073/pnas.0700930104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lund K, Liu Y, Lindsay S, Yan H. Self-assembling a molecular pegboard. J Am Chem Soc. 2005;127:17606–17607. doi: 10.1021/ja0568446. [DOI] [PubMed] [Google Scholar]
- 36.Park SH, Pistol C, Ahn SJ, Reif JH, Lebeck AR, Dwyer C, LaBean TH. Finite-size, fully addressable DNA tile lattices formed by hierarchical assembly procedures. Angew Chem Int Ed. 2006;45:735–739. doi: 10.1002/anie.200503797. [DOI] [PubMed] [Google Scholar]
- 37.Liu Y, Ke Y, Yan H. Self-assembly of symmetric finite-size DNA nanoarrays. J Am Chem Soc. 2005;127:17140–17141. doi: 10.1021/ja055614o. [DOI] [PubMed] [Google Scholar]
- 38.Chworos A, Severcan I, Koyfman AY, Weinkam P, Oroudjev E, Hansma HG, Jaeger L. Building programmable jigsaw puzzles with RNA. Science. 2004;306:2068–2072. doi: 10.1126/science.1104686. [DOI] [PubMed] [Google Scholar]
- 39.Rothemund PWK, Papadakis N, Winfree E. Algorithmic self-assembly of DNA Sierpinski triangles. PLoS Biol. 2004;2:2041–2053. doi: 10.1371/journal.pbio.0020424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barish RD, Rothemund PWK, Winfree E. Two computational primitives for algorithmic self-assembly: copying and counting. Nano Lett. 2005;5:2586–2592. doi: 10.1021/nl052038l. [DOI] [PubMed] [Google Scholar]
- 41.Chen H-J, Schulman R, Goel A, Winfree E. Reducing facet nucleation during algorithmic self-assembly. Nano Lett. 2007;7:2913–2919. doi: 10.1021/nl070793o. [DOI] [PubMed] [Google Scholar]
- 42.Fujibayashi K, Hariadi R, Park SH, Winfree E, Murata S. Toward reliable algorithmic self-assembly of DNA tiles: a fixed-with cellular automaton pattern. Nano Lett. 2008;8:1791–1797. doi: 10.1021/nl0722830. [DOI] [PubMed] [Google Scholar]
- 43.Schulman R, Winfree E. Synthesis of crystals with a programmable kinetic barrier to nucleation. Proc Natl Acad Sci USA. 2007;104:15236–15241. doi: 10.1073/pnas.0701467104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen J, Seeman NC. The synthesis from DNA of a molecule with the connectivity of a cube. Nature. 1991;350:631–633. doi: 10.1038/350631a0. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y, Seeman NC. The Construction of a DNA Truncated Octahedron. J Am Chem Soc. 1994;116:1661–1669. [Google Scholar]
- 46.Goodman RP, Schaap IAT, Tardin CF, Erben CM, Berry RM, Schmidt CF, Turberfield AJ. Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication. Science. 2005;310:1661–1665. doi: 10.1126/science.1120367. [DOI] [PubMed] [Google Scholar]
- 47.Goodman RP, Berry RM, Turberfield AJ. The single-step synthesis of a DNA tetrahedron. Chem Commun. 2004:1372–1373. doi: 10.1039/b402293a. [DOI] [PubMed] [Google Scholar]
- 48.Goodman RP, Heilemann M, Doose S, Erben CM, Kapanidis AN, Turberfield AJ. Reconfigurable, braced, three-dimensional DNA nanostructures. Nat Nanotechnol. 2008;3:93–96. doi: 10.1038/nnano.2008.3. [DOI] [PubMed] [Google Scholar]
- 49.Erben CM, Goodman RP, Turberfield AJ. Single-molecule protein encapsulation in a rigid DNA cage. Angew Chem Int Ed. 2006;45:7414–7417. doi: 10.1002/anie.200603392. [DOI] [PubMed] [Google Scholar]
- 50.Shen Z, Yan H, Wang T, Seeman NC. Paranemic crossover DNA: a generalized Holliday structure with applications in nanotechnology. J Am Chem Soc. 2004;126:1666–1674. doi: 10.1021/ja038381e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aldaye FA, Sleiman HF. Modular access to structurally switchable 3D discrete DNA assemblies. J Am Chem Soc. 2007;129:13376–13377. doi: 10.1021/ja075966q. [DOI] [PubMed] [Google Scholar]
- 52.He Y, Ye T, Su M, Zhang C, Ribbe AE, Jiang W, Mao C. Hierarchical self-assembly of DNA into symmetric supramolecular polyhedral. Nature. 2008;452:198–202. doi: 10.1038/nature06597. [DOI] [PubMed] [Google Scholar]
- 53.Zhang C, Su M, He Y, Zhao X, Fang P, Ribbe AE, Jiang W, Mao C. Conformational flexibility facilitates self-assembly of complex DNA nanostructures. Proc Natl Acad Sci USA. 2008;105:10665–10669. doi: 10.1073/pnas.0803841105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Le JD, Pinto Y, Seeman NC, Musier-Forsyth K, Taton TA, Kiehl RA. DNA-templated self-assembly of metallic nanocomponent arrays on a surface. Nano Lett. 2004;4:2343–2347. doi: 10.1021/nl0515495. [DOI] [PubMed] [Google Scholar]
- 55.Zhang J, Liu Y, Ke Y, Yan H. Periodic square-like gold nanoparticle arrays template by self-assembled 2D DNA nanogrids on a surface. Nano Lett. 2006;6:248–251. doi: 10.1021/nl052210l. [DOI] [PubMed] [Google Scholar]
- 56.Sharma J, Chhabra R, Liu Y, Ke Y, Yan H. DNA-templated self-assembly of two-dimensional and periodical gold nanoparticle arrays. Angew Chem Int Ed. 2006;45:730–735. doi: 10.1002/anie.200503208. [DOI] [PubMed] [Google Scholar]
- 57.Zheng J, Constantinou PE, Micheel C, Alivisatos AP, Kiehl RA, Seeman NC. Two-dimensional nanoparticle arrays show the organizational power of robust DNA motifs. Nano Lett. 2006;6:1502–1504. doi: 10.1021/nl060994c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sharma J, Chhabra R, Cheng A, Brownell J, Liu Y, Yan H. Control of self-assembly of DNA tubules through integration of gold nanoparticles. Science. 2009;323:112–116. doi: 10.1126/science.1165831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin C, Ke Y, Liu Y, Mertig M, Gu J, Yan H. Functional DNA nanotube arrays: bottom-up meets top-down. Angew Chem Int Ed. 2007;46:6089–6092. doi: 10.1002/anie.200701767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sharma J, Ke Y, Lin C, Chhabra R, Wang Q, Nangreave J, Liu Y, Yan H. DNA-tile-directed self-assembly of quantum dots into two-dimensional nanopatterns. Angew Chem Int Ed. 2008;47:5157–5159. doi: 10.1002/anie.200801485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aldaye FA, Sleiman HF. Sequential self-assembly of a DNA hexagon as a template for the organization of gold nanoparticles. Angew Chem Int Ed. 2006;45:2204–2209. doi: 10.1002/anie.200502481. [DOI] [PubMed] [Google Scholar]
- 62.Sharma J, Chhabra R, Anderson C, Gothelf K, Yan H, Liu Y. Toward reliable gold nanoparticle patterning on self-assembled DNA nanoscaffold. J Am Chem Soc. 2008;130:7820–7821. doi: 10.1021/ja802853r. [DOI] [PubMed] [Google Scholar]
- 63.Park SH, Yin P, Liu Y, Reif JH, LaBean TH, Yan H. Programmable DNA self-assemblies for nanoscale organization of ligands and proteins. Nano Lett. 2005;5:729–733. doi: 10.1021/nl050175c. [DOI] [PubMed] [Google Scholar]
- 64.Liu Y, Lin C, Li H, Yan H. Aptamer directed self-assembly of proteins on a DNA nanostructure. Angew Chem Int Ed. 2005;44:4333–4338. doi: 10.1002/anie.200501089. [DOI] [PubMed] [Google Scholar]
- 65.Chhabra R, Sharma J, Ke Y, Liu Y, Rinker S, Lindsay S, Yan H. Spatially addressable multiprotein nanoarrays template by aptamer-tagged DNA nanoarchitectures. J Am Chem Soc. 2007;129:10304–10305. doi: 10.1021/ja072410u. [DOI] [PubMed] [Google Scholar]
- 66.Ke Y, Lindsay S, Chang Y, Liu Y, Yan H. Self-assembled water-soluble nucleic acid probe tiles for label-free RNA hybridization assays. Science. 2008;319:180–183. doi: 10.1126/science.1150082. [DOI] [PubMed] [Google Scholar]
- 67.Lin C, Liu Y, Yan H. Self-assembled combinatorial encoding nanoarrays for multiplexed biosensing. Nano Lett. 2007;7:507–512. doi: 10.1021/nl062998n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rinker S, Ke Y, Liu Y, Chhabra R, Yan H. Self-assembled DNA nanostructures for distance-dependent multivalent ligand-protein binding. Nat Nanotechnol. 2008;3:418–422. doi: 10.1038/nnano.2008.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Duckworth BP, Chen Y, Wollack JW, Sham Y, Mueller JD, Taton TA, Distefano MD. A universal method for the preparation of covalent protein-DNA conjugates for use in creating protein nanostructures. Angew Chem Int Ed. 2007;46:8819–8822. doi: 10.1002/anie.200701942. [DOI] [PubMed] [Google Scholar]
- 70.Malo J, Mitchell JC, Vénien-Bryan C, Harris JR, Wille H, Sherratt DJ, Turberfield AJ. Engineering a 2D protein-DNA crystal. Angew Chem Int Ed. 2005;44:3057–3061. doi: 10.1002/anie.200463027. [DOI] [PubMed] [Google Scholar]
- 71.Seeman NC. The construction of 3-D stick figures from branched DNA. DNA Cell Biol. 1991;10:475–486. doi: 10.1089/dna.1991.10.475. [DOI] [PubMed] [Google Scholar]
- 72.Eckardt LH, Naumann K, Matthias Pankau W, Rein M, Schweitzer M, Windhab N, Von Kiedrowski G. DNA nanotechnology: Chemical copying of connectivity. Nature. 2002;420:286. doi: 10.1038/420286a. [DOI] [PubMed] [Google Scholar]
- 73.Lin C, Xie M, Chen JJL, Liu Y, Yan H. Rolling-circle amplification of a DNA nanojunction. Angew Chem Int Ed. 2006;45:7537–7539. doi: 10.1002/anie.200602113. [DOI] [PubMed] [Google Scholar]
- 74.Lin C, Wang X, Liu Y, Seeman NC, Yan H. Rolling circle enzymatic replication of a complex multi-crossover DNA nanostructure. J Am Chem Soc. 2007;129:14475–14481. doi: 10.1021/ja0760980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lin C, Rinker S, Wang X, Liu Y, Seeman NC, Yan H. In vivo cloning of artificial DNA nanostructures. Proc Natl Acad Sci USA. 2008;105:17626–17631. doi: 10.1073/pnas.0805416105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liedl T, Sobey TL, Simmel FC. DNA based nano-devices. Nanotoday. 2007;2:36–41. [Google Scholar]
- 77.Seeman NC. From genes to machines: DNA nanomechanical devices. Trends Biochem Sci. 2005;30:119–125. doi: 10.1016/j.tibs.2005.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bath J, Turberfield AJ. DNA nanomachines. Nat Nanotechnol. 2007;2:275–284. doi: 10.1038/nnano.2007.104. [DOI] [PubMed] [Google Scholar]
- 79.Salaita K, Wang Y, Mirkin CA. Applications of dip-pen nanolithography. Nat Nanotechnol. 2007;2:145–155. doi: 10.1038/nnano.2007.39. [DOI] [PubMed] [Google Scholar]
- 80.Huang Y, Duan X, Wei Q, Lieber CM. Directed assembly of one-dimensional nanostructures into functional networks. Science. 2001;291:630–633. doi: 10.1126/science.291.5504.630. [DOI] [PubMed] [Google Scholar]
- 81.Zhang RS, McCullum EO, Chaput JC. Synthesis of two mirror image 4-helix junctions derived from glycerol nucleic acid. J Am Chem Soc. 2008;130:5846–5847. doi: 10.1021/ja800079j. [DOI] [PubMed] [Google Scholar]
- 82.Lin C, Ke Y, Li Z, Wang JH, Liu Y, Yan H. Mirror image DNA nanostructures for chiral supramolecular assemblies. Nano Lett. 2009;9:433–436. doi: 10.1021/nl803328v. [DOI] [PMC free article] [PubMed] [Google Scholar]