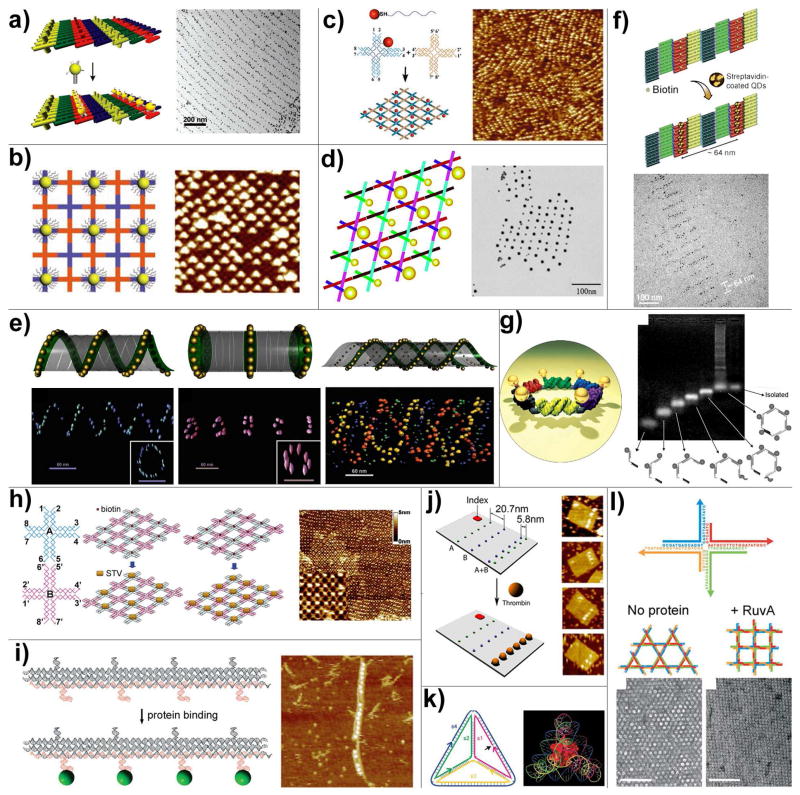
Figure 5.
DNA directed assembly of multi-component nanoarrays. (a) Organization of 5 nm AuNPs on DNA DX lattices. (b) Periodic 5 nm AuNP nanoarrays with well controlled interparticle distances templated by 2D DNA nanogrids. (c) DNA mono-modified 5 nm AuNPs directly participate in the self-assembly process and yield periodic nanoparticle arrays. (d) 2D periodic array of 5- and 10-nm AuNPs generated by incorporating DNA mono-modified AuNPs into robust triangle-shaped DNA motifs. (e) Controlled self-assembly of DNA tubules through integration of AuNPs. The assembly results in 3D nanoparticle architectures such as single-spiral tube (left), stacking ring tube (middle) and interlocking double-spiral tube (right). The schematic views are placed above corresponding electron tomographic images. (f) Quantum dots organized on DNA DX lattices through biotin-streptavidin interaction. (g) Discrete hexagonal AuNP array displayed on a DNA hexagon consisting of six non-identical molecules each with two ssDNA arms linked by an organic molecule. (h) Programmable streptavidin 2D arrays formed on biotinylated DNA lattices. (i) 1D thrombin array assembled by incorporating anti-thrombin aptamers into linear TX DNA array. (j) Selective binding of thrombin proteins to the bivalent anti-thrombin aptamers displayed on the surface of rectangular origami arrays. (k) A cytochrome c protein trapped inside a DNA tetrahedron cage. (l) The binding of RuvA to Holliday junction tiles alters the assembly product from Kagome-type lattice (left) to square-planar lattice (right).