Figure 1. Structures of the Ca-ATPase and PLB.
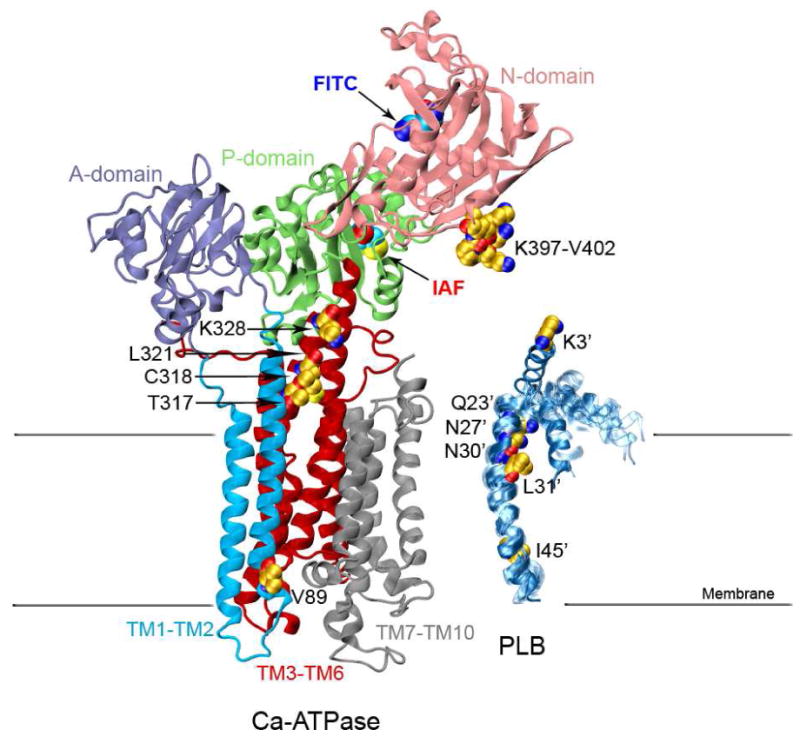
Structure of the Ca-ATPase (1su4.pdb) depicts headpiece, corresponding to discrete structural elements associated with the A-domain (residues 1-43 and 124-235; purple), N-domain (residues 360-600; pink), and P-domain (residues 330-359 and 601-739; green) relative to the transmembrane helices TM1-TM2 (residues 44-123; cyan), TM3-TM6 (residues 239-329 and 740-821, red), and TM7-TM10 (residues 831-994; gray), where domain boundaries are as previously described (72). The ten NMR structures of a PLB monomer are shown on the right (1fjk.pdb). Highlighted in the structures of Ca-ATPase and PLB are (yellow space-filling) side chains that have been identified through covalent cross-linking experiments which involve both endogenous amino acids (C or K) or engineered cysteines at sites in TM2 (i.e., V89), TM4 (T317, C318, L321, and K328), and the N-domain (i.e., K397DDKPV402) of the Ca-ATPase with sites on PLB (i.e., I45-L52, L31, N30, N27, Q23, or K3) (9-11). Labeling sites are indicated for FITC modification at K514 /K515 in the N-domain (blue oval) and IAF modification at C674 in the P-domain (red oval) of the Ca-ATPase.
