Figure 8. Proposed Structural Model for the Ca-ATPase Homodimer.
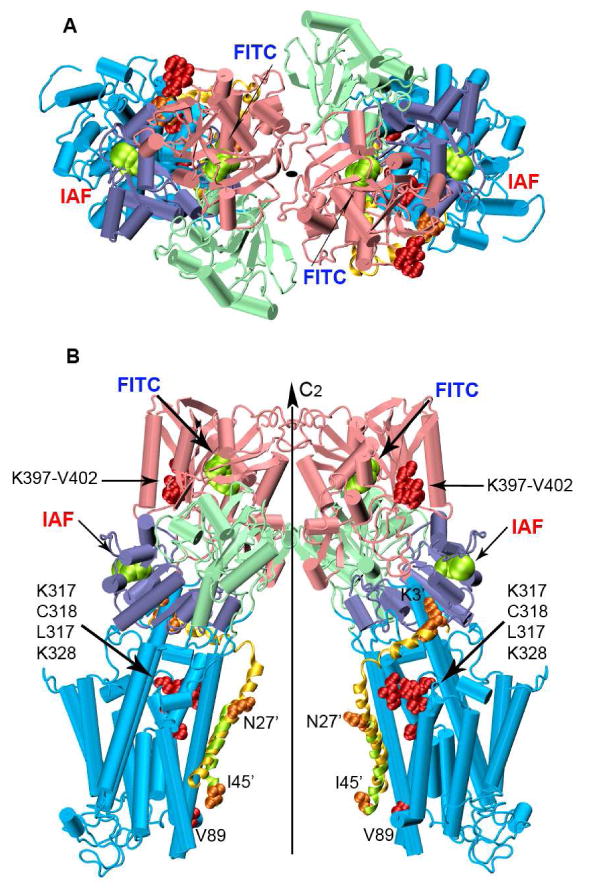
The two monomers of the Ca-ATPase are related by a two-fold rotational center of symmetry perpendicular to the cytoplasmic membrane. Top (A) and longitudinal (B) views of the dimer built from the crystallographic structure of SERCA1, i.e., 1kju.pdb. Domains are colored as in Figure 1. The lowest energy PLB conformer from the NMR ensemble (1fjk.pdb) was superimposed on the lowest energy docked PLB conformer (domain Ia and II; Figure S3), and does not consider known large-scale changes in the orientation of the N-domain of the Ca-ATPase upon PLB binding (10, 56, 67). The NMR-derived and the predicted PLB conformers are shown in yellow and green cartoon helices respectively. Amino acid residues involved in PLB-Ca-ATPase crosslinking are represented as spacefilling structures: orange for PLB and red for the Ca-ATPase. PLB residues have been marked with the prime symbol.
