Table 1. Crystal data and refinement statistics.
Values in parentheses are for the highest resolution shell.
| Crystal data | |
| Space group | Monoclinic P21 |
| Unit-cell parameters (Å, °) | a = 42.3, b = 41.2, c = 72.3, β = 104.1 |
| Molecules per ASU | 1 |
| VM (Å3 Da−1) | 2.09 |
| Solvent content (%) | 41.2 |
| Data collection | |
| Resolution (Å) | 23.4–1.1 (1.14–1.1) |
| Unique reflections | 92539 |
| No. of frames | 300 |
| Total No. of reflections | 162663 |
| Rmerge† (%) | 6.8 (39.0) |
| I/σ(I) | 13.8 (3.3) |
| Completeness (%) | 94.7 (96.0) |
| Mean multiplicity | 2.9 (2.5) |
| Refinement statistics | |
| No. of reflections used | 87911 |
| Rwork/Rfree‡ (%) | 11.2/14.7 |
| Reflections used for Rfree (|Fo| > 0) (%) | 5.0 |
| R.m.s. deviation, bonds (Å) | 0.014 |
| R.m.s. deviation, 1–3 distances (Å) | 0.030 |
| Ramachandran plot (%) | |
| Most favored | 89.8 |
| Allowed | 9.7 |
| Generously allowed | 0.5 |
| Model | |
| Protein atoms including alternate conformations | 2100 |
| Zn2+ | 1 |
| Glycerol (1) | 6 |
| AZM (3) | 39 |
| Waters, full/half occupancy | 348/56 |
| Average temperature factors (Å2) | |
| Main chain | 11.3 |
| Side chain | 15.7 |
| Waters (fully occupied only) | 32.0 |
| AZM 701/702/703 | 11.6/15.9/21.6 |
R
merge = 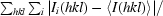
 × 100, where Ii(hkl) is the intensity of an individual reflection and 〈I(hkl)〉 is the average intensity.
× 100, where Ii(hkl) is the intensity of an individual reflection and 〈I(hkl)〉 is the average intensity.
R
work = 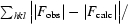
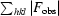 × 100; R
free is identical to R
work but for 5% of data omitted from refinement.
× 100; R
free is identical to R
work but for 5% of data omitted from refinement.
