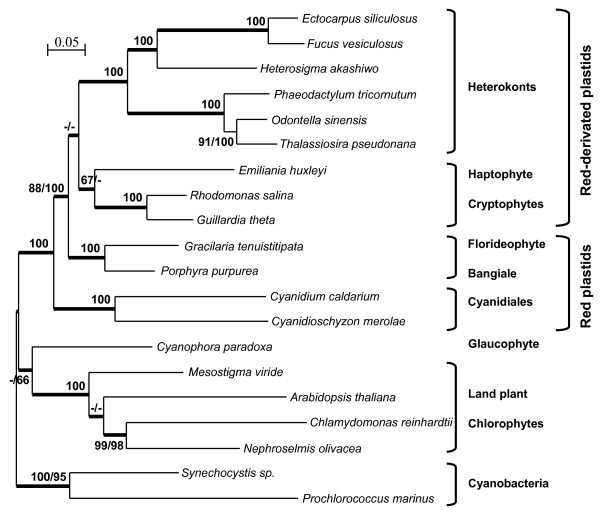
Figure 3.
Maximum likelihood tree constructed from a dataset of 44 concatenated proteins from 20 plastid or cyanobacterial complete genomes. PHYML and Neighbour Joining trees were constructed based on 8,652 amino-acid sites using cpREV and JTT matrices, respectively. When above 65% and different, bootstrap values (1000 replicates) are provided for PHYML (first value) and NJ (second value) analyses. The thick branches represent ≥ 0.9 posterior probability for Bayesian inference analysis.