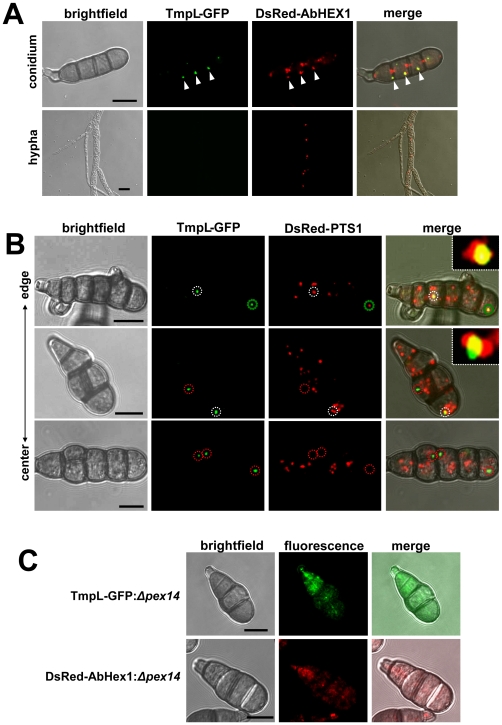
Figure 2. Subcellular distribution of TmpL-GFP fusion protein.
(A) Co-localization analyses with a TmpL-GFP and DsRed-AbHex1 double-labeled strain in an A. brassicicola conidium (upper panel) and vegetative hypha (lower panel) were examined using confocal microscopy. TmpL-GFP localizes over cell cortex-associated or cytoplasmic DsRed-AbHex1 signals (arrowheads), not the septal pore-associated signals. Note that no TmpL-GFP signal is observed in the growing vegetative hyphae. Bars = 10 µm. (B) Co-localization analyses with a TmpL-GFP and DsRed-PTS1 double-labeled strain in A. brassicicola conidia. DsRed-PTS1 fluorescence reveals peroxisomes. A large peroxisome is completely associated with TmpL-GFP signal (a green dotted circle, top panel). The other circles denote a partial association between TmpL-GFP and DsRed-PTS1 signals (white dotted circles, top and middle panels) and a complete dissociation of TmpL-GFP with DsRed-PTS1 signals (red dotted circles, middle and bottom panels). Note that different conidial age determined by collected sites, from the center to the edge of fungal colony, shows different types of association between two fluorescence signals. Insets indicate a magnified view of each white dotted circle representing a partial association between TmpL-GFP and DsRed-PTS1 signals. Bars = 10 µm. (C) Localization analyses with Δpex14 mutants on a background of either a TmpL-GFP or DsRed-AbHex1 strain. Note that the deletion of pex14 resulted in cytoplasmic redistribution of TmpL-GFP and DsRed-AbHex1 fluorescence signals. Bars = 10 µm.