Abstract
The catalytic activity of the histone deacetylase (HDAC) enzymes is directly relevant to the pathogenesis of cancer as well as several other diseases. HDAC inhibitors have been shown to have the potential to treat several types of cancers. The role of computational study of the HDAC enzymes is reviewed, with particular emphasis on the important role of molecular modeling to the development of HDAC inhibitors with improved efficacy and selectivity. The use of two computational approaches—one structure-based, and the second ligand-based—toward inhibitors against the different HDAC sub-classes, are summarized.
INTRODUCTION
The opposing catalytic activities of the histone acetyltransferases (HATs) and histone deacetylases (HDACs) are critical in the regulation of gene transcription [1]. The HATs and HDACs are two distinct families used for the reversible change of the chemical state of ε-amino group of the lysine residues residing at the N-termini of the core histone proteins. The chemical state of these lysines (whether as the free ε-amine or the N-acetylated ε-amine) profoundly effects chromatin remodeling and the epigenetic regulation of genes [2]. Acetylation is one of the most abundant epigenetic modifications of the histones, and acetylation positively correlates with active gene expression. Conversely, deacetylation is positively correlated to gene silencing.
Aberrant control of the state of histone acetylation/deacetylation leads to the development of several types of cancers [3]. HDAC inhibition leads to a hyperacetylated state of the histones, initiating the transcriptional activation of suppressed genes. In cancer states hyperacetylation ultimately causes cell cycle arrest and apoptosis. This approach recently was proven to be a valid strategy for the treatment of cancer [4]. The first HDAC inhibitor, SAHA (Vorinostat®), was approved in 2006 by the FDA for the chemotherapy of cutaneous T-cell lymphoma (CTCL). Several other very promising HDAC inhibitors are currently in Phase I/II clinical trials [5]. Accumulated evidence also shows that HDAC inhibition may have therapeutic benefit against several central nervous system disorders [6] and against malaria [7].
Human HDACs are grouped into four classes (Classes I, II, III and IV) on the basis of sequence homology to their yeast orthologues (Rpd3, Hda1, and Sir2) [8], Table 1. Classes I, II and IV are Zn(II)-dependent HDACs, containing 11 separate enzymes denoted as HDACs 1-11. The Class III HDACs are NAD+-dependent, and having a catalytic mechanism that is distinctly different from the other three HDAC classes. Class I HDACs (HDAC1, 2, and 3), each about 350–500 amino acid residues in length, are usually recruited as subunits of transcriptional multiprotein complexes in the nucleus. For example, HDAC1 and 2 are involved in the NuRD and Sin3 repressor complexes [9]. HDAC3 is part of the N-COR and SMRT complexes [10]. Class II is divided into two sub-classes, the class IIa enzymes (HDAC4, 5, 6, 7, and 9) and the class IIb enzymes (HDAC6, 10). Class IIa HDACs have distinct tissue-specific patterns of expression, and are predominantly localized in muscle and heart tissues [11]. The class IIa HDACs comprises a total of 500-600 residues, reflecting the presence of an extended C-terminal domain not found in the class I HDACs. This domain plays a key role in the interaction of the class IIa HDACs with other proteins, including HDAC3 and the myocyte enhancer factor 2 (MEF2). HDAC6, one of the class IIb HDACs, is unique among the entire HDAC family. It contains two independent catalytic domains and a zinc finger ubiquitin-binding domain at its C-terminus. HDAC6 catalyzes deacetylation of acetylated α-tubulin, thus altering the stability of the microtubules in cytoplasm [12]. HDAC10, a second class IIb member, lacks the second catalytic domain found in HDAC6. Its function is still unknown. HDAC11 is the only member of class IV, and is primarily localized in the nucleus. Little information is available regarding its expression and function. Sequence alignment implies that its structure may be related to both classes I and II HDACs.
Table 1.
Characteristics of Human HDACs and related X-ray structures
| Class | Member | Sequence Sizea |
UnitProtKB | Cellular Localization |
Related PDB codeb |
|---|---|---|---|---|---|
| HDAC1 | 482 | Q13547 | Nucleus | NAc | |
| I | HDAC2 | 488 | Q92769 | Nucleus | NAd |
| HDAC3 | 428 | O15379 | Nucleus | NAd | |
| HDAC8 | 377 | Q9BY41 | Nucleus | 1T64, 1T67, 1T69, 1VKG[19a]; 1W22[19b]; 2V5W, 2V5X[21] |
|
| IIa | HDAC4 | 1084 | P56524 | Nucleus/Cytoplasm | 2VQJ, 2VQQ, 2VQM, 2VQO, 2VQV[22] |
| HDCA5 | 1122 | Q9UQL6 | Nucleus/Cytoplasm | NAc | |
| HDAC7 | 952 | Q8WUI4 | Nucleus/Cytoplasm | 3C0Y,3C0Z,3C10 [23] | |
| HDAC9 | 1011 | Q9UKV0 | Nucleus/Cytoplasm | NAc | |
| IIb | HDAC6 | 1215 | Q9UBN7 | Cytoplasm | NAe |
| HDAC10 | 669 | Q969S8 | Cytoplasm | NAc | |
| IV | HDAC11 | 347 | Q96DB2 | Nucleus/Cytoplasm | NAc |
| SIRT1 | 747 | Q96EB6 | Nucleus | NAc | |
| SIRT2 | 389 | Q8IXJ6 | Cytoplasm | 1J8F[25]f | |
| SIRT3 | 399 | Q9NTG7 | Mitochondria | NAc | |
| III | SIRT4 | 314 | Q9Y6E7 | Mitochondria | NAc |
| SIRT5 | 310 | Q9NXA8 | Mitochondria | 2NYR, 2B4Y[26] | |
| SIRT6 | 355 | Q8N6T7 | Nucleus | NAc | |
| SIRT7 | 400 | Q9NRC8 | Nucleus | NAc |
Sequence lengths of human HDACs are from http://www.uniprot.org/uniprot/
Catalytic domain only.
Not Available, but bacterial homologue structures 1C3P, 13CR and 1C3S [16] are available.
Not Available.
Not Available, but bacterial HDHA structures 1ZZ0, 1ZZ1, 1ZZ3 and 2GH6 [24] are available.
The Sirtuin, or class III HDACs, includes seven enzymes (SIRT1 to 7). Each shows substantial sequence similarity to the yeast protein Sir2 (silent information regulator 2) [13]. These HDACs use NAD+ as a co-substrate during their catalysis of hydrolytic deacetylation. Each sirtuin has a conserved catalytic domain, comprising 270 amino acid residues. Human SIRT1 regulates energy metabolism, aging and the circadian rhythm [14]. SIRT1 specifically targets acetylated lysines at position 3 of Histone1 (H1), position 16 of Histone3 (H3), and position 14 of Histone4 (H4). Acetylated lysines in non-histone proteins (such as p53, TAFI68, and p300) are also SIRT1 substrates. Like HDAC-6, SIRT-2 also can remove an acetyl group from acetylated α-tubulin and microtubules, as well as at lysine16 of H4. SIRT3 to 5 are characterized as mitochondrial sirtuins. There is little information about their targets and function.
The biological function and relevance of the HDAC enzymes, as well as the design and synthesis of their inhibitors, have been reviewed regularly [15-17]. In contrast, the computational study of these important pharmaceutical targets is much less frequently summarized. Here, an overview is provided of the recent computational work on these important enzymes. This review is organized as follows. First, a description of current computational techniques for protein ligand studies is given, followed by structural features of HDACs with practical applications concerning docking, homology modeling, molecular dynamics (MD) simulation. QSAR (Quantitative structure-activity relationship) and other computational methods for the purpose of identifying inhibitors against the HDAC sub-classes follow. Finally, prospective directions of this field are discussed.
CURRENT COMPUTATIONAL APPROACHES FOR ENZYME INHIBITOR DESIGN
Computer-aided molecular design is an integral aspect of drug design and discovery. In general, computational methods for drug discovery and design may be divided into two categories. The first category is structure-based drug design. Using available 3D structural and other important biological information concerning the target protein, the binding strength of small molecule inhibitors is optimized. The 3D structural information is obtained either from experiment (X-ray or NMR determination of the protein structure) or from reliable computational methods (such as homology modeling, using a known structure showing high sequence similarity to the target protein). Molecular mechanical (MM) as well as quantum mechanical (QM) approaches may be used to identify small molecules with binding affinity, and then to refine the structure of these small molecules toward higher binding affinity.
Quite often, the 3D structural information of the target protein is not available and there is no good template for homology modeling. In this case, a second category of computational techniques, ligand based methods such as QSAR are used. From the dataset obtained from a series of lead compounds, 2D or 3D descriptors are generated. QSAR equations are derived based on the 2D descriptors, and usually a pharmacophore model is created from the 3D descriptors. The QSAR equation and the pharmacophore model are used to suggest new compounds with improved activity.
STRUCTURAL FEATURES OF HDACS
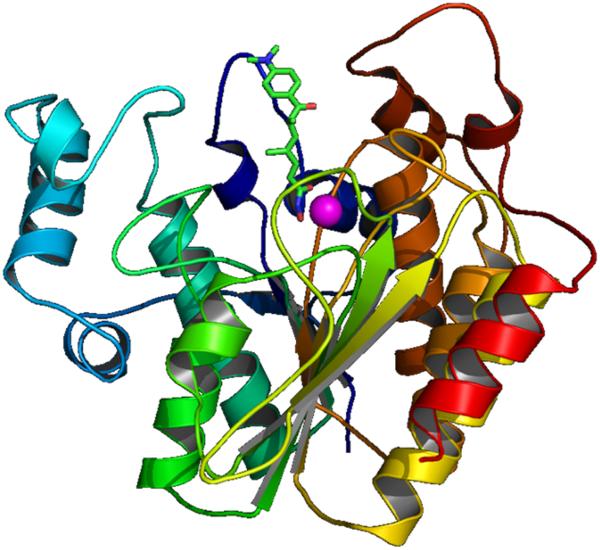
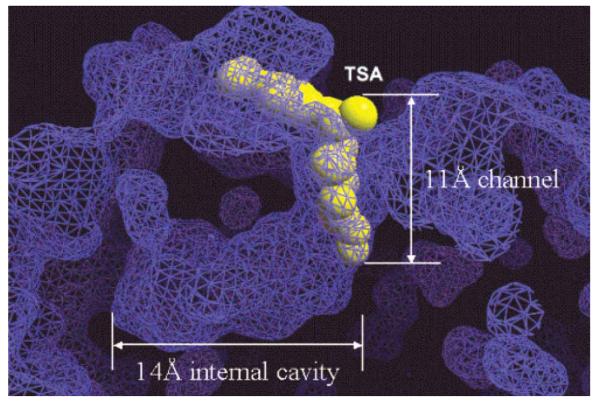
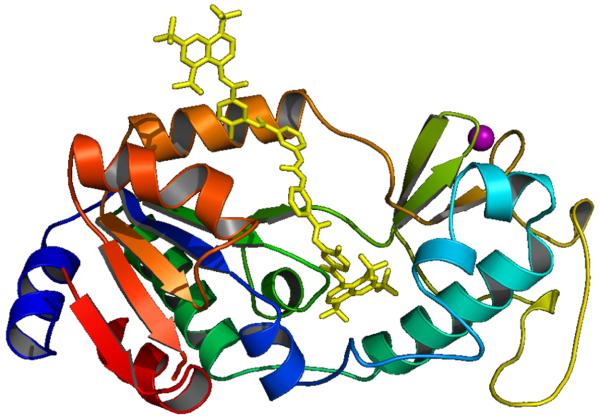
The first X-ray structure (2.0 Å resolution) of an HDAC-related protein, the histone deacetylase-like protein (HDLP) from Aquifex aeolicus, in complex with two HDAC inhibitors (TSA and SAHA) was elucidated by Finnin et al. in 1999 [18]. These X-ray structures revealed a tube-like, 11 Å deep channel for accommodation of the TSA and SAHA structure (Fig. 1). The long chain found in the lysine substrate as well as the inhibitors TSA and SAHA makes several favorable contacts with the hydrophobic residues of the protein lining this channel. The hydroxamic acid group in TSA and SAHA binds to the active site Zn(II) center, located at the bottom of this channel. Just adjacent to the channel, there is a 14 Å deep internal cavity formed by Arg, Tyr and Cys residues (Chart 3 and Fig. 6). This internal cavity is thought to be a common structural feature of the class I HDACs, at least for the HDAC1, HDAC2 and HDAC3 isoforms which have high sequence similarity.
Fig. (1).
TSA (green stick) in the active site of HDLP. Zn: magenta ball.
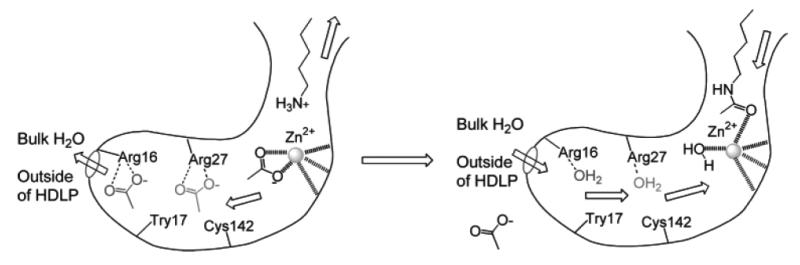
Chart 3.
Proposed function of 14 Å internal cavity for transferring deacetylation byproduct and water molecules. Image reprinted with permission from [20].
Fig. (6).
Surface representation of 11 Å channel and 14 Å internal cavity of HDLP. TSA is displayed as a yellow CPK model. Image reprinted with permission from [20].
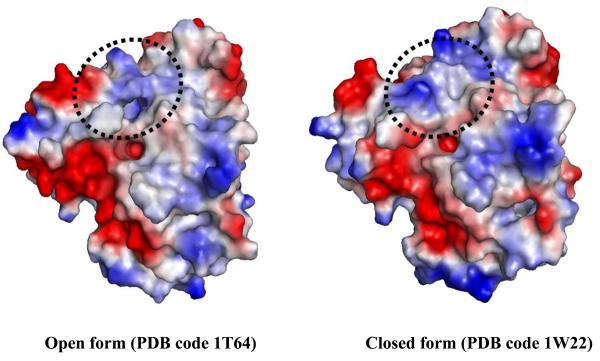
The sequence of HDAC8 is unique compared to other members of the class I HDACs [8]. Nonetheless, the two X-ray structures of human HDAC8, which were solved by two groups, show a substrate entry channel [19] corresponding to the 11 Å channel found in HDLP. The internal cavity adjacent to the tunnel is also present. However, the dimensions for this cavity found in the two structures are different [19], suggesting that the cavity size may be dynamic (Fig. 2). Comparing the two structures, significantly different conformations for the Ser30-Lys36 loop, located between the last and second-to-last helices, are observed. To some extent this difference supports an earlier hypothesis that the function of 14 Å internal cavity is to accommodate, and ultimately to facilitate, the egress of the acetate reaction by-product to bulk solvent [18, 20]. The critical role of Asp101 in substrate binding and interaction with inhibitors was confirmed by lowered enzymatic activity and in binding assays of Asp101 mutants of HDAC8 [21].
Fig. (2).
Surface representation of open and closed forms of the second pocket, corresponding to 14 Å internal cavity, in human HDAC8. The area with major structural change is indicated by the dotted circle.
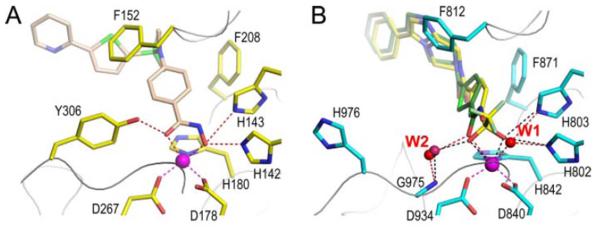
Two X-ray structures of the catalytic domain of the human class IIa enzymes HDAC7 and HDAC4 recently were solved [22-23]. The overall fold of both HDAC7 and HDAC4 is similar to previously reported HDAC structures. The principle difference is in a loop region around the active site entrance. Both HDAC7 and HDAC4 have a second, surface exposed zinc binding motif adjacent to the exit of the active site that is not observed in class I HDACs. This motif very likely participates in substrate recognition and protein-protein interactions. A more subtle difference is that in the class I/IIb HDAC structures obtained thus far, the hydroxyl group of a conserved tyrosine (Tyr306 of HDAC8 or Tyr312 of HDAH—Histone Deacetylase-like AmidoHydrolase) that is suggested to provide a stabilizing hydrogen bond to the oxyanion intermediate encountered during catalysis (Fig. 3) is replaced by a histidine conserved in all class IIa HDACs. In HDAC4, this position is occupied by a histidine (His976) whose side chain points away from the active site and does not bind to a ligand. A water molecule (W2) bridges between Glu975 and the oxygen atom in either the carbonyl of the substrate or the hydroxamate inhibitors (Fig. 3). If the function of the tyrosine is to stabilize the oxanion of the intermediate, it can be hypothesized that the class IIa enzyme may have lower enzymatic activity due to loss of this interaction [22].
Fig. (3).
The active sites of HDAC8 (A, 1W22, inhibitor: light brown carbon) and HDAC4 (B: 2VQQ, TMFK: yellow carbon; 2VQM, HA: green carbon). Image reprinted with permission from [23].
Only one class IIb related structure is available, that of the C-terminal portion of the bacterial protein HDAH isolated from Bordetella/Alcaligenes strain FB188 [24]. The folding of HDAH is very similar to that seen for the class I HDAC structures, except for the two different loops. The first loop, which is not present in class I HDACs, contains residues 16 to 38 and may be class IIb-specific. The second loop, including residues 92-96, unfortunately, has a poorly defined structure. Among eukaryotic HDACs, the catalytic domains of human HDAC6 show the highest sequence homology to FB188 HDAH. Therefore, HDAH is the best template available for homology modeling of the catalytic domains of HDAC6.
A number of sirtuin structures are known at this time. These structures include two human sirtuins, SIRT2 [25] and SIRT5 [26], and several SIRT2 homologues from other species [27-31, 69-70]. The overall domain organization and folds are very similar among all of these structures. A large domain (approximately 180 residues) having a Rossmann fold is seen, similar to what is found in many NADH/NADPH-binding enzymes. A smaller domain contains a structural Zn binding motif (Fig. 4). NAD+ binds in a pocket between the two domains. Larger structural variations among all SIRT structures are seen in the amino- and carboxyl-terminal extensions. In the Rossmann fold domain, a longer helix and a loop insertion is observed for SIRT2 than is observed for SIRT5. A unique inserted loop in the structural zinc binding motif is found in the SIRT5 structure compared to the other sirtuin structures.
Fig. (4).
X-ray structure of human SIRT5 with bounded suramin (yellow stick). Large subdomain with Rossmann fold (Left). Structural Zn containg domain (right). Zn is shown as magenta ball. Only chain A in X-ray structure (PDB code 2NYR) is displayed here.
STRUCTURE-BASED STUDY OF HDACS
Class I HDACs
Among all HDAC isoforms, HDAC1 is the one most studied since its activity most directly correlates to human diseases such as cancer. Numerous HDAC1 inhibitors have been discovered in the last ten years, many as a result of structure-based design using HDAC structure and computer modeling of HDAC-inhibitor interactions. Since no structure of human HDAC1 has been solved so far, several homology models of human HDAC1 have been constructed [20, 32-33]. These models have enriched our understanding at the atomic level as to how HDAC inhibitors interact with HDAC1, and why some inhibitors have stronger inhibition activity.
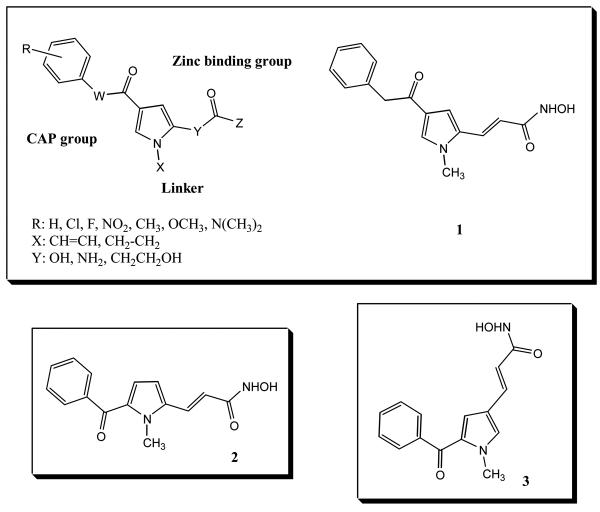
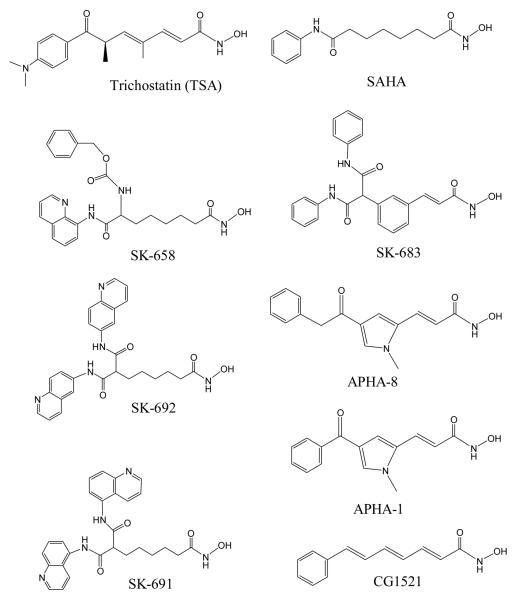
Massa et al. [33] reported a series of aroyl-pyrrolyl-hydroxyamides (APHAs) with low micromolar activity against the maize histone deacetylase HD2 (Chart 1). An initial docking study showed that the pyrrole was a linker, interacting with the tube-like 11 Å substrate entry channel. Optimization of the activity of this inhibitor class was done by modification at C-2 and/or C-4 of the pyrrole, and by alteration of the number of methylene units in the linker region [34]. Using a human HDAC1 homology model, three docking approaches were compared. Initial binding mode analyses were carried out by a semiautomatic dock (SAD) method. Two docking studies were performed in parallel, using DOCK 4.02 and AutoDock 3.0.5 software. Three bound APHA conformations were obtained, and ranked with their in-house Validate scoring function.
Chart 1.
APHA HDAC inhibitors.
Three more potent APHA molecules were identified computationally, and confirmed by synthesis and assay as HDAC inhibitors. Compound 1 gave IC50 values of 0.1 μM against maize HD2 and 0.5 μM against mouse HDAC1. Compound 2 had very similar activities against these two enzymes (for both enzymes, IC50 values of 0.3 μM). Compound 3 is a stronger HD2 inhibitor (IC50 value of 0.05 μM) than mouse HDAC1 (0.78 μM). Docking results showed different orientations of the pyrrole linkers. In these docking studies, compounds 2 and 3 gave a better fit into the model of the HDAC1 binding cavity than 1 (Fig. 5). The hydroxyamate groups in 2 and 3 are closer to the Zn(II) ion. The (C)O---Zn and (H)O---Zn distances are respectively 3.3 Å, 2.7 Å for 2; 2.9 Å, 3.2 Å for 3 and 4.0 Å, 4.7 Å for 1. The docking studies suggest three positive interactions to account for the tight fit of 3 in the homology model of HDAC1: (1) favorable π-π stacking interactions between the pyrrolyl-ethylene and the Phe140 and Phe197 residues; (2) a hydrogen bond network among the hydoxamate group, His130, His131 and Asp167 sidechains and the catalytic water; (3) a hydrogen bond between Tyr296 and the oxygen atom of the hydroxamate group. It is worth mentioning that the N-methylpyrrole in 3 does not make crucial interactions with any residues in HDAC1. This may allow the propenoylhydroxamate tail to better reach the Zn(II) ion. A slightly different picture was observed for the complex of 2. The N-methyl group of 2 contacts Gly139, and the benzoyl oxygen atom may interact with Tyr90 through a water-bridged hydrogen bond. These additional interactions were suggested [34] to contribute to the stronger inhibition activity of 2.
Fig. (5).
AutoDock binding mode of 2 (right, shown as ball-and-stick) and 3 (left, shown as ball-and-stick). Image reprinted with permission from [34].
In the meantime, we carried out a systematic docking study of nine strong HDAC1 inhibitors into the HDLP structure (Chart 2). We focused on identifying common features of inhibitor binding to HDLP/HDAC1 and analyzed HDAC structure to see if other structural features beyond the 11 Å channel in HDAC are important for inhibitor binding [20]. We treated the side chains of His131 and His132 as neutral state with protonated Nδ atom. The hydroxymate group in the docked ligands were treated as neutral as well. The docking poses for TSA with HDLP showed excellent agreement to the X-ray structure. The RMSD of all of the heavy atoms of TSA was 0.9 Å. In the X-ray structure, the Zn2+⋯O=C and Zn2+⋯O(H) distances were 2.45 Å and 2.24 Å respectively, and 2.54 Å and 1.84 Å in the best docked structure. This close result are in line with improved density function theory calculations for hydroxamic acid and hydroxamates bound to a simple HDAC active site model [35, 36]. Other calculations using different model systems and may be different protonation states gave poorer results (2.9–4.0 Å, 3.2–4.7 Å) [34]. The docking result for the HDLP-SAHA complex also agreed well with the X-ray structure. The successful outcomes with these two inhibitors give confidence that the calculated HDLP complexes with SK-683, APHA-8, SK-658, SK-692, and CG1521 are reasonable models [20].
Chart 2.
Known HDAC inhibitors included in our study [20].
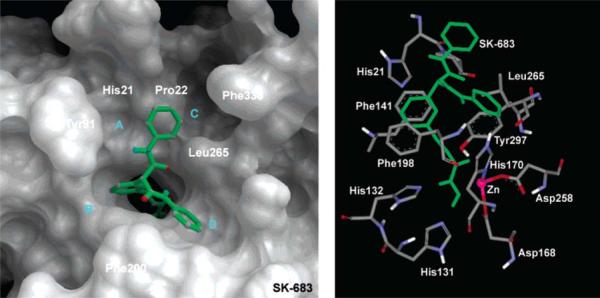
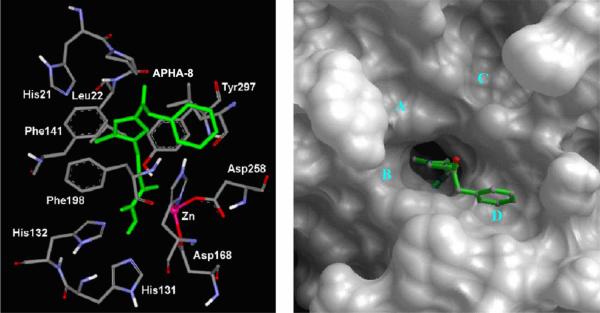
Several general and important features for those complexes were found. (1) Four shallow pockets (A to D) on the surface of the active site exit in HDLP perfectly accommodate aromatic groups in the “cap region” (i.e. the surface binding group found in most HDAC inhibitors). In particular, these interactions may explain the pM potency of SK-683 as an HDAC inhibitor. The aromatic group of the SK-683 linker makes favorable π-π interactions with the two parallel Phe141 and Phe198 residues, similar to the methyl group of TSA (Fig. 7). (2) The APHA-8 complex suggests the N1-methyl of APHA-8 is positioned similarly to the C-4-methyl group of TSA, as seen in the X-ray structure. The N1-methyl forms favorable CH⋯π interaction with Phe141 and Phe198. The aromatic group at C-4 of the pyrrole occupies pocket D on the surface of HDLP (Fig. 8). These inhibitor conformation and location are different to what was calculated by others [34]. (3) The 14 Å internal cavity is proposed as a pipeline to transfer the acetate by-product of the hydrolysis from the active site away from the catalytic center (Chart 3 and Fig. 6). This result suggests that this cavity can serve as an additional pocket for occupancy by substituents of HDAC inhibitors. Although these conclusions were drawn from the HDLP system, it is reasonable to postulate that the complexes with human HDAC1 are similar [37]. This hypothesis was substantiated subsequently by others [38]. Moradei et al. demonstrated that the 14 Å internal cavity indeed can be used as a second binding pocket for additional interactions. Based on this hypothesis, aminophenyl benzamide-type HDAC inhibitors with enhanced potency and selectivity were designed and synthesized. These inhibitors target primarily HDAC1 and 2, with weaker activity against HDAC3 and 11[38].
Fig. (7).
View of surface and active site of SK-683 bound to HDLP. Image reprinted with permission from [20].
Fig. (8).
View of surface and active site of APHA-8 bound to HDLP. Image reprinted with permission from [20].
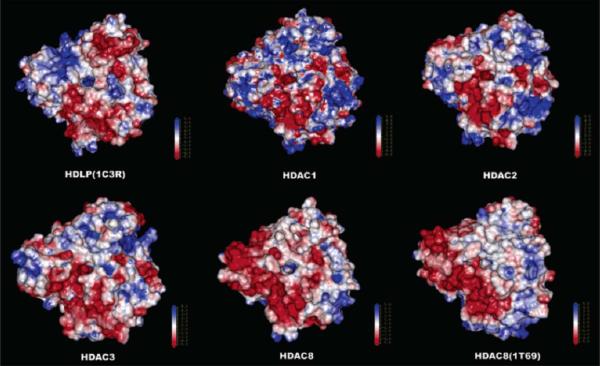
We further developed four human class I HDAC homology models for HDACs1, 2, 3 and 8 [37]. Six HDAC inhibitors were docked into each active site. From this study, it was concluded that the 11 Å channel is difficult to exploit for the design of isoform-selective HDAC inhibitors. Instead, the subtle differences in the shape and charge distribution around the entrance of the 11 Å channel is potentially a better means of differentiating among these four HDACs (Fig. 9). As the differences between HDAC1 and 2 around the entrance are very small, the design of an inhibitor to selectively distinguish between HDAC1 and 2 is anticipated to be very difficult. MD simulations of several HDAC-inhibitor complexes suggest that the conformational behavior of the cap group of inhibitor is important to binding affinity [37]. Finally, a comparison between our HDAC8 homology model and the X-ray structure showed excellent agreement and thus validated our homology modeling [37].
Fig. (9).
Electrostatic surfaces on a scale of +5 to −5 for homology models and X-ray structures of HDLP and HDACs 1, 2, 3, and 8. Image reprinted with permission from [37].
While most groups used nonbonded Zn(II) model for modeling of HDAC-inhibitor interactions, Park and Lee used bonded Zn(II) parameters to simulate the complexes of several inhibitors with a homology model of DAC1. Free energy perturbation methods were applied to estimate the relative free energy of binding of nine HDAC inhibitors through fairly short MD trajectory (300 ps). The relative calculated free energy calculated agreed well with the order of potency obtained from experimental observation [32].
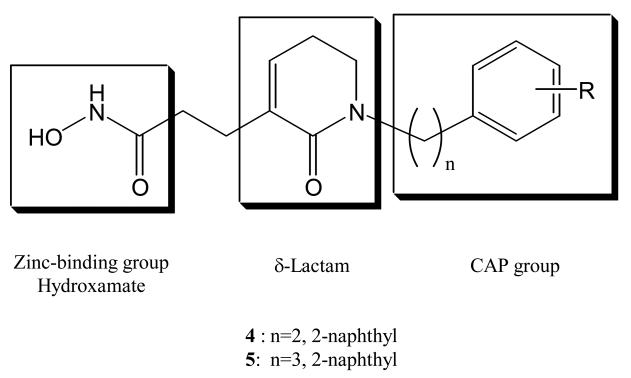
Kim et al. designed several δ-lactam-based HDAC inhibitors incorporating modified cap groups [39]. The objective was to incorporate a δ-lactam linker domain between the surface recognition cap group and the zinc-binding hydroxamate. Compounds 4 and 5 (Chart 4) were the most potent inhibitors from among the twenty compounds studied. The respective IC50 values for these two compounds (37 nM and 30 nM) against HDAC1 are approximately four-fold more potent than SAHA. Compound 4 showed the most potent growth inhibitory activity against five human tumor cell lines (PC-3, ACHN, NUGC-3, HCT-15, and MBA-MB-231). In order to understand the binding mode for these inhibitors, a docking study based on the crystal structure of HDLP (PDB code 1C3R) was done. A model of the catalytic core of human HDAC1 was made using the Discover program. The hydroxamic acid moiety of 4 chelates to the catalytic Zn(II) ion, and the δ-lactam interacts with the tubular hydrophobic pocket. In addition, the hydrophobic 2-naphthyl cap group forms favorable interactions with the protein surface. Their docking result also suggested that the chain length between the hydrophobic cap group and the δ-lactam, and the size of cap group, are both very critical for formation of the flexible conformation required to occupy the binding pocket of human HDAC1. These findings are consistent with the previous studies [20, 37].
Chart 4.
δ-Lactam based HDAC inhibitors
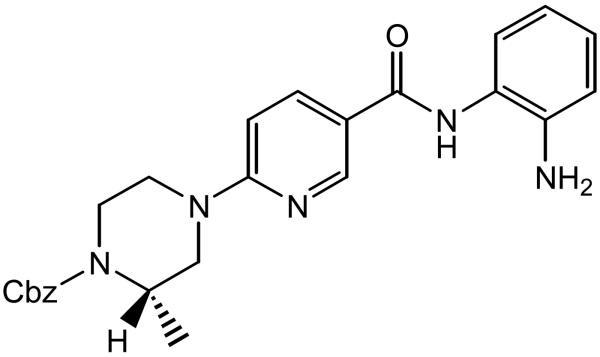
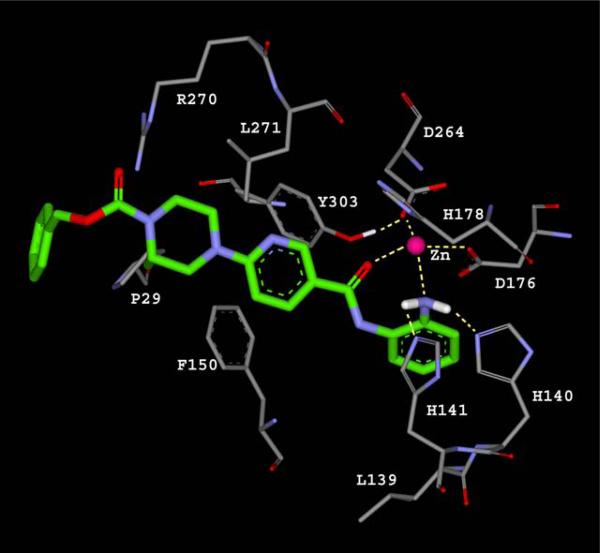
Further effort toward an HDAC inhibitor with improved class I selectivity was reported by Hamblett et al. [40], using MS-275 as the lead compound and with structurally diversification around its pyridine ring. A human homology model based on the combination of two X-ray structures—HDLP (PDB code 1C3S) and human HDAC8 (PDB code 1T64)—was created to understand the interactions of inhibitor 6 (Chart 5). The NH2 group of the aniline, and the carbonyl oxygen of the nicotinamide, were assumed to bind to the Zn(II) ion through a 7-membered ring chelate. The amino group of the aniline also forms hydrogen bonds with His140 and His141, while the nitrogen lone pair of the NH2 interacts with the Zn(II) ion 2.4 Å away. This distance is close to the 2.2 Å distance obtained from density functional theory calculations [35, 36]. The minimized structure showed the piperazine in a chair-like conformation, allowing the benzyl carbamate to efficiently interact with the protein surface. The side chain of Phe150 rotates by almost 90° to form an edge-to-face π stacking interaction with the amide NH (Fig. 10). The observed selectivity of 6 for HDAC1 over HDAC8 was suggested to arise from exclusion of the aniline ring from the channel in HDAC8 by the bulky side chain of Trp141, while the Leu139 at the same position in HDAC1 allows such binding.
Chart 5.
6-piperazinyl nicotinamides 6 as HDAC inhibitors
Fig. (10).
A proposed binding mode for bezamide inhibitor (green) in human HDAC1 homology model. Image redrew from [40].
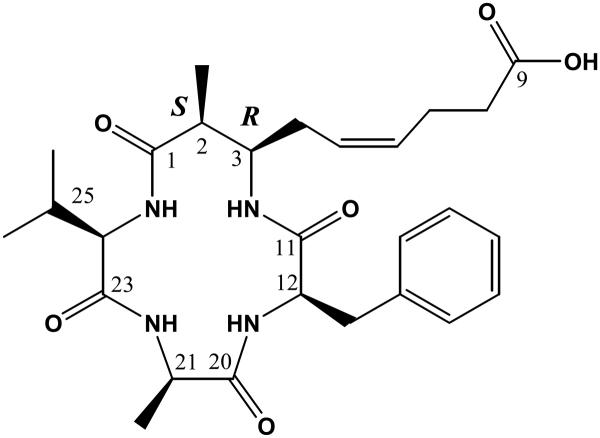
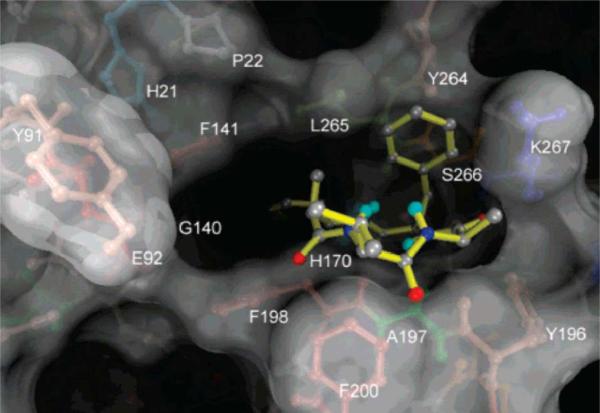
Azumamides A–E were isolated recently from the marine sponge Mycale izuensis [41]. Structurally, they have three d-α-amino acids and a unique β-amino acid. Biological assay undertaken by Maulucci et al. [42] indicated that Azumamide E (Chart 6) was a selective inhibitor of HDAC1, 2 and 3, with respective IC50 values of 50 nM, 100 nM and 80 nM. It is one of the few highly potent and selective HDAC inhibitors having a carboxylate as the zinc binding group. The calculated complex for azumamide E and HDLP is shown in Fig. 11. The d-Phe side chain is nicely surrounded by a pocket defined by the Tyr264, Leu265, Ser266 and Arg267 residues. The cyclotetrapeptide core interacts with the shallow pocket on the receptor, which we have denoted as pocket D [20]. The long aliphatic chain bearing a carboxylate group inserts into the 11 Å channel to allow carboxylate binding to the Zn(II) ion .
Chart 6.
Azumamide E as class I selective HDAC inhibitor
Fig. (11).
Docked azumamide E (yellow) in the active site of HDLP. O: red; N: dark blue; C: gray; Polar H: cyan. Image reprinted with permission from [42].
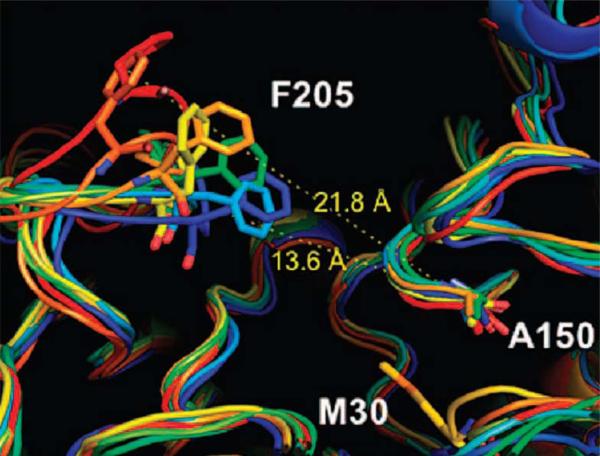
Weerasinghe et al. investigated the influence of residues in the 11 Å channel of HDAC1 on its enzymatic activity by alanine scanning mutagenesis [43]. The residues in the 11 Å channel were found to be critical for deacetylase activity. Eight alanine mutants (H28A, P29A, D99A, G149A, F150A, Y204A, F205A and L271A) showed a 62-91% reduction in activity compared to wild-type enzyme. Nonetheless, a 2 ns MD simulation suggested that the channel residue mutants maintained the overall wild-type structure, and that the global protein structure was not altered significantly. In the native enzyme, Y204 and F205 display significant rotations prior to ligand binding. The F150A mutant showed significantly greater flexibility at F205 position. Because a favorable interaction exists between these two phenyl rings in the native structure, it is possible that the loss of this interaction causes movement of F205 (Fig. 12). Interestingly, the structurally similar F150Y mutant is inactive. This dramatic loss of activity in this mutant may result from a hydrogen bond between the hydroxyl group of Y150 and D99. Such an interaction changed the side chain conformation of D99 and opened the channel for F205 to undergo a flip. F205 and Y150 would not be parallel anymore. Therefore, the structure of this changed channel may not be as good as that in the wild-type enzyme for inhibitor binding.
Fig. (12).
F150A mutant shows increased flexibility of the F205 containing loop. (Blue: first snapshot; red: last snapshot). Image reprinted with permission from [43].
Class II HDACs
Considerable effort has been directed toward isoform-selective HDAC inhibitor design, as discussed in several well-prepared reviews [44-45]. Since the active site residues for all HDAC isoforms are almost identical, isoform selectivity remains a quite challenging objective. Only a few class II-selective HDAC inhibitors (HDAC4 or 6) have been reported [46-48].
Schafer et al. reported biaryalaine-containing hydroxamic acids as class IIb-selective HDAC6 inhibitors, with modest selectivity for HDAC6 over HDAC1 (Chart 7) [49]. Since HDAC6 has two catalytic domains, it is unclear that which one (or whether both) contribute to catalytic activity. Two conflicting interpretations have been argued. Zhang et al. suggest that both domains should be equally active in HDAC6 [50], while Zou et al. showed that the C-terminal domain is the major functional one [51]. To address the question of whether both domains function equally, both catalytic domains of HDAC6 were generated using homology modeling using three X-ray structures of HDAC8 (PDB code 1T64), HDAC7 (PDB code 2PQO) and HDLP (PDB code 1C3R) as homology templates. The two catalytic domain models show high structural similarity. Large differences are observed at the entrance of the 11 Å channel. The docking study demonstrated large variations in HDAC-inhibitor interactions on the cap region of compound 7 for HDAC6 catalytic domain I and catalytic domain II, while the rest of compound 7 interacts similarly with both catalytic domains. Only catalytic domain II of HDAC6 explains the experimentally-derived structure-activity relationships. This result supports the conclusion that the second HDAC-6 domain is the predominant binding site for HDAC-6 inhibitors [51].
Chart 7.
Class IIb selective HDAC6 inhibitors. Compound 7: n=6
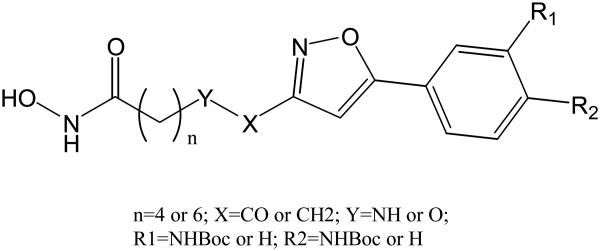
A set of phenylisoxazole-containing hydroxamates (Chart 8) was reported to have HDAC6 selectivity with picomolar activity [52]. Compound 8 shows good HDAC6 selectivity. A homology model of HDAC6 was built using the X-ray structure of HDAC7 [52]. A docking study using 8 revealed that its hydroxamic acid group and six methylene linker bind deep inside the tube-like channel. An interaction between carbonyl group of the Boc moiety and His499 was found. This may help the cap region of 8 interacting with the protein surface (Fig. 13). Since there is no such an interaction for the other HDAC inhibtiors in the same study, this may explain the high activity of 8 against HDAC6. Among various classes HDACs, significant differences in the loop region adjacent to the active site channel which interacts with the cap group were found. These differences may account for the observed high selectivity.
Chart 8.
HDAC6 selective phenylisoxazole-containing HDAC inhibitors. Compound 8: R1=H, R2=NHBoc, n=6
Fig. (13).
Compound 8 docked into the active site of HDAC6 model. Image reprinted with permission from [52].
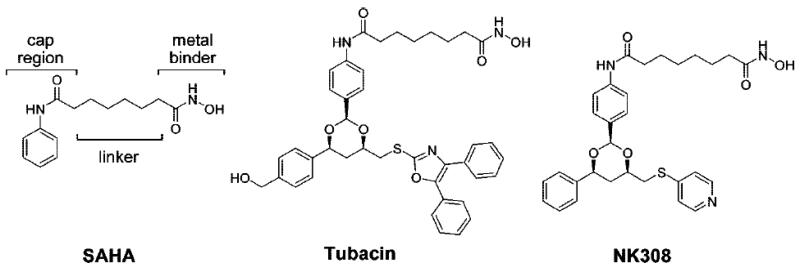
Estiu et al. [53] performed molecular dynamics simulations to study the structural origin of selectivity of the class II-selective HDAC inhibitors SAHA, tubacin and NK308 (Chart 9). A homology model of human HDAC6 with only one HDAC domain was created using the structure of the bacterial HDAC6 homologue FB188 HDAH (PDB code 1ZZ1). Sequence alignment showed 38% similarity between human HDAC6 and FB188 HDAH. The model was refined through consecutive MD minimizations. In order to see low frequency motions, trajectories of at least 10 ns were calculated using Amber 8 and ff02 force field parameters. Enzyme assays showed that SAHA is an almost equally strong inhibitor for HDAC1 and HDAC6 (48 nM and 21 nM respectively), but it is a weak inhibitor for HDAC8 (~2000 nM). In contract, tubacin and NK308 are more potent inhibitors of HDAC6 (142 nM and 28 nM respectively) than HDAC1 (995 nM and 88 nM).
Chart 9.
HDAC inhibitors studied with 10-ns MD simulation [53].
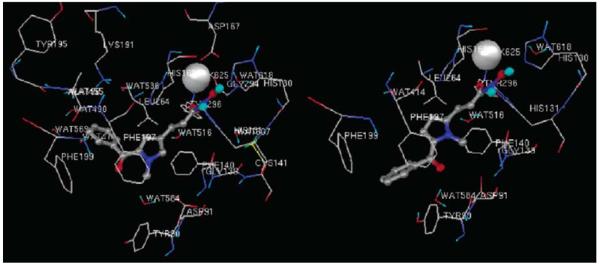
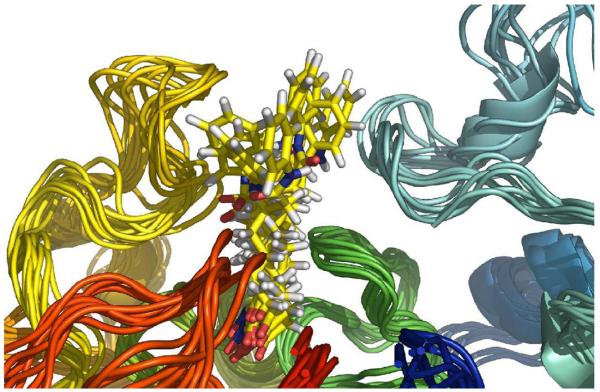
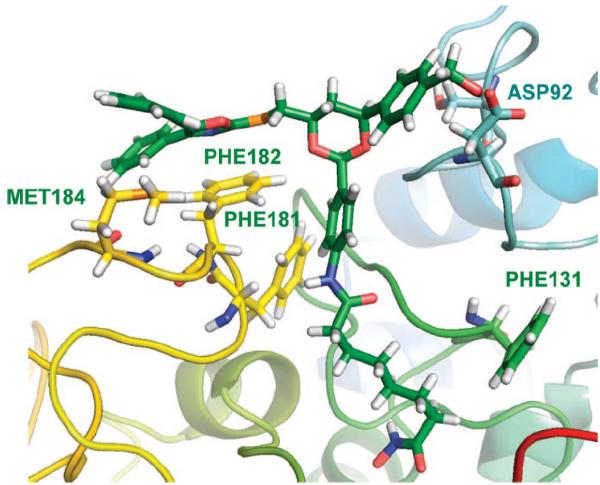
Trajectory snapshots plotted from the simulation indicated that there is no preferred interaction between the cap group in SAHA and the surface of HDAC1. Rather, SAHA reached different pockets on the surface with equal probability. Similar results were obtained for the complexes of SAHA with HDAC6 and 8 (Fig. 14). On the other hand, tubacin showed quite different binding modes for HDACs 1, 6 and 8. In HDAC6·tubacin complex, the 2,3-diphenyloxazole is close to the Phe181 and Phe182 residues at the mouth of the active site (Fig. 15). Tyr201 and Tyr204 of HDAC1 also participate in tubacin binding in the HDAC1-tubacin complex but this interaction in turn changed the position of hydroxymethylphenyl moiety to a solvent-exposed position. Such a binding mode difference would explain the lower binding affinity of tubacin to HDAC1 compared to HDAC6. Thus, the different structural characteristics and shape of the protein surface around the mouth of the active site, as well as the differences in protein flexibility between HDAC6 and HDAC8, are important to the future rational design of class II- vs. class I-selective HDAC inhibitors.
Fig. (14).
Snapshots from the MD simulation of SAHA in HDAC6 show no preferred protein-CAP group interactions. Image reprinted with permission from [53].
Fig. (15).
Phe181 and 182 interact with 2,3-diphenyl oxazole in HDAC6-tubacin complex in MD simulation. Image reprinted with permission from [53].
Class III NAD+-dependent HDACs
The study of class III NAD+-dependent HDACs has been bloomed in recent years. Of the seven human SIRTs, SIRT1 has been the most extensively studied since it is highly expressed in several adult tissues, such as those of the brain, heart, and in skeletal muscle [54]. It also interacts with several important transcriptional factors, such as p53 and NF-kB to regulate their activity [67]. A recent review on class III HDAC inhibitors was published [56]. Two recent computational studies related to this enzyme are summarized here.
Huhtiniemi et al. published a comparative modeling of human SIRT1 [57]. In order to investigate the active site-ligand interactions and design novel SIRT1 inhibitors, a model of the catalytic core domain (residues 244 to 498) of the full length of SIRT1 was built using the HOMOLOGY module implemented in Insight II. Six X-ray structures of the sirtuin decaetylase family (Human SIRT2, PDB code 1J8F [25]; Saccharomyces cerevisiae HST2, PDB code 1Q1A [27]; Thermotoga maritima SIR2Tm, PDB code 1YC5 [28]; Archaeoglobus fulgidus SIR2-Af2, PDB code 1S7G [29]; Archaeoglobus fulgidus SIR2-Af1, PDB code 1M2G [30] and Eschericia Coli. cobB, PDB code 1S5P [31]) were used as templates. The model was minimized with the CVFF force field in Insight II to remove the bad contacts and further refined with MD simulation using the GROMACS package to locate favorable side chain conformations. S and R enantiomers of 19 indole derivates (38 total structures) were docked into the binding site of the SIRT1 model using the GOLD. The best pose was selected according to GoldScore fitness function. The results demonstrated that an hydrogen bond network among the amide group of the indole in EX-527, shown in Chart 10 and D348, T349, I347, N346 as well as Q345 is well established in the C-pocket, which is part of the flexible loop (from 269 to 295) with highly conserved residues in the sirtuin family.
Chart 10.
Structure of EX-527 (S-isomer)
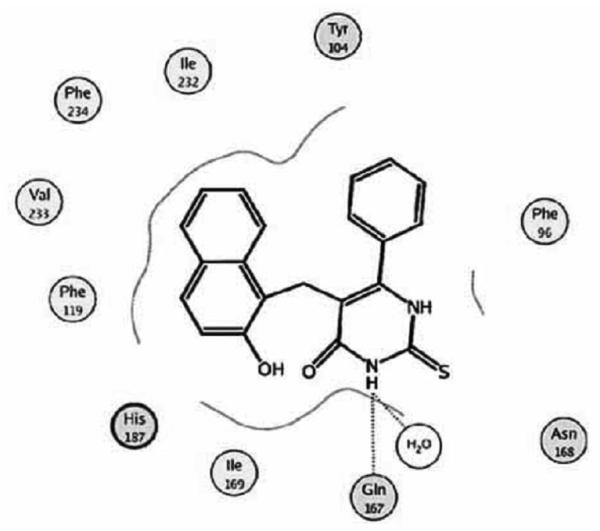
Neugebauer et al. reported a docking study for various SIRT2 inhibitors against the human SIRT2 X-ray structure using the GOLD [58]. Their initial docking result showed that cambinol interacts with the nicotinamide subpocket and the lysine-substrate channel (Chart 11). This is in agreement with the experimental observation [59]. Consideration of four water molecules near the active site significantly improved the docking result. Residues Gln167 and Asn 168 form hydrogen bonds with a polar group in cambinol.
Chart 11.
Docked cambinol in the active site of human SIRT2 model. Image reprinted with permission from [58].
LIGAND-BASED STUDY OF HDACS
Although 3D structural information for HDACs and their inhibitors is available, it is still quite challenging to identify potent compounds from a huge compound pool. Docking of every compound from the pool is not practical. What structural descriptors are significant to active compound selection? A ligand-based approach may give an answer. At least, this approach helps to decrease the size of the candidate compound pool, by quickly eliminating unlikly ones and further increases the selection ratio (active vs. inactive). In addition, this approach is much faster than routine docking calculations and can include physicochemical parameters that allow the incorporation of ADME properties in whole-cell studies. This will make it quite suitable to apply to huge (size of millions) compound pool. Selected QSAR and 3D-QSAR studies for HDACs are presented here.
QSAR Studies
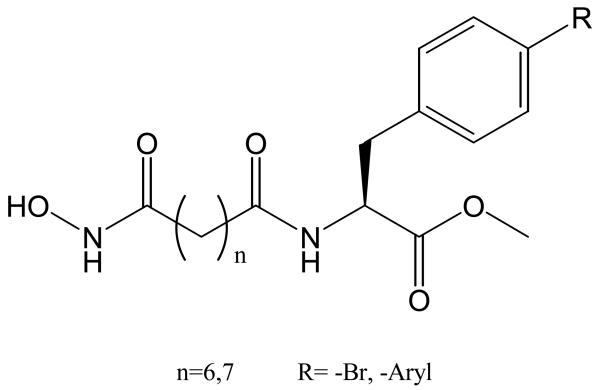
Using calculated physiochemical descriptors of nineteen TSA and SAHA-like hydroxamic acids [60], we derived two QSAR equations for PC-3 cell line inhibition activity [61]. Both equations gave the same physical meaning with three variables. Further analysis of the two equations indicated that Eq. 1 was more reliable than Eq. 2.
| (Eq. 1) |
| (Eq. 2) |
Eq. 1 clearly indicates that a high charge reflecting a more polarized C=O bond in the N-hydroxyl-amide group indicates a compound will have strong inhibition activity. The negative coefficient globularity term Glob means a compound with a lower Glob value will have stronger inhibiton activity. This is consistent with the fact that a linear shape compound with a straight linker 6 or 7 carbons long is usually a good HDAC inhibitior.
Xie et al. did a QSAR study on histone deacetylase inhibitors [62] in 2004. They analyzed and reconstructed a fairly large biological activity dataset with 124 compounds from various published sources which covered most important HDAC inhibitors available at that time. The dataset was further reorganized as dataset1 (47 compounds, most of which are TSA/SAHA-type linear hydroxamic acids, several bearing a sulfonamide group), dataset2 (62 compounds, more diverse than dataset1), and dataset3 (94 compounds). Finally, three QSAR equations, Eq. 3-5, were built with four descriptors, a_aro, vsa_hyd, SlogP_VSA9 and PEOE_VSA+0. The most significant descriptors in these models are vsa_hyd (approximation to the sum of VDW surface of hydrophobic atoms of enzyme-binding region of the HDAC inhibitor), and SlogP_VSA9 (corresponding to atomic contribution of logP (oct/ow) > 0.40). A_aro and PEOP_VSA are related to the number of aromatic atoms and the partial charge of certain atoms.
For dataset1
| (Eq. 3) |
For dataset2
| (Eq. 4) |
For dataset3
| (Eq. 5) |
Kozikowski et al. designed and synthesized a series of substituted biaryl hydroxamates and mercaptoacetamides as HDAC inhibitors against pancreatic cancer cell growth with nanomolar potency [63]. Five QSAR equations (Eq. 6-10) were developed from the 23 compounds (biphenyl or phenylthiazoles bearing hydroxamates or mercaptoacetamides) against HDACs 1, 2, 8, 10 and 6 incorporating the binary indicators I-NHCOCH2SH and I-Thiazole and calculated LogP (ClogP). If the compound is mercaptoacetamide, then I-NHCOCH2SH = 1.0; otherwise it is 0. If the compound is phenylthiazole, then I-Thiazole = 1.0; otherwise it is 0.
| (Eq. 6) |
| (Eq. 7) |
| (Eq. 8) |
| (Eq. 9) |
| (Eq. 10) |
It is clear from these results that the inhibition activities against different isoforms of HDAC are highly correlated. For instance, Eq 6 and Eq 9 are very similar although they are developed for the HDAC1 and HDAC7 isoforms. This again demonstrates the subtleties necessary to obtain isoform selective HDAC inhibitors.
3D QSAR Studies
Usually, 3D QSAR studies are accompanied by docking studies. So far, only a few applications have been published for class I HDAC inhibitors [64-67] and only one for class II HDAC inhibitor [68]. To my knowledge, no 3D QSAR study for class III HDAC inhibitors has been published to date.
Chen et al. developed a 3D QSAR pharmacophore model from 30 known HDAC inhibitors to identify essential ligand features for HDAC inhibition activity [55]. The model, Hypo1, includes HBA (hydrogen bond acceptor) and HBD (hydrogen bond donor) features corresponding to the metal-binding function and representing coordination to the Zn(II) ion. In addition, hydrophobic/π-π stacking interaction between the ligand and enzyme also play a critical role for the inhibition activity. A comparison of TSA conformation between in Hypo1 and in the HDLP-TSA complex showed a RMSD of 1.19 Å. Then 25 known HDAC inhibitors were applied to such a model to validate its prediction ability. The predicted and experimental activities showed a fairly good correlation coefficient (0.896), confirming the validity of the model.
Ragno et al. [68] carried out a 3D QSAR study for their new developed class II-selective HDAC inhibitors (APHAs) against maize HD1-A and HD1-B with modest selectivity. 25 APHAs were used in their training set. Conformational search by simulated annealing in continuum solvent simulation (GBSA) was applied to these molecules. The optimized structures were imported by Almond software (MIA, Perugia, Italy) to generate Grind descriptors. It was found that the most important variable is the distance between an H-acceptor bond group (C=O) and an aromatic portion of the cap (DRY-N1: 6 for HD1-B and DRY-N1: 7 for HD1-A). The models further suggest that a bent molecular shape is important for selective HD1-A activity, while a straight molecule is good for HD1-B activity. 14 compounds were used to validate the prediction power of the models for class I/II selectivity. Both models correctly predicted that all compounds in the test set were class II-selective HDAC inhibitors.
SUMMARY AND PERSPECTIVES
This review presents an overview of the computational studies used to identify novel and potent HDAC. Thanks to the elucidation of HDAC X-ray structures and the study of molecular modeling, many active HDAC inhibitors have been discovered. Several very promising compounds have undergone human clinical trials. Molecular modeling has shown its ability as a useful tool to develop novel HDAC inhibitors. The valuable information obtained from molecular modeling has enriched our understanding of these biologically important targets.
Several important issues should be considered in the future study: 1) Regulating the level of different HDACs has significant biological impact. Among the HDAC inhibitors that have been studied so far, there are only a few isoform-selective inhibitors. Even rarer are compounds that have high selectivity. Therefore, the development of novel inhibitors with higher isoform-selectivity is needed. Computational studies will continue to have significant impact toward this objective. 2) The majority of HDAC inhibitors have a hydroxamic acid zinc-binding group. Because of the concerns regarding toxicity of hydroxamic acid, the identification of alternative zinc-binding groups within the HDAC inhibitors is highly desirable. 3) In most computational studies of HDAC enzymes, the ionization states of the HDAC enzymes and the bound ligands were not discussed or compared. Therefore, it is highly recommended to consider this important issue later. 4) All published studies used docking methods that treated the HDAC enzyme as a rigid structure. The flexibility of the HDAC8 second pocket and surface mobility demonstrated in the few X-ray structures demands future consideration of protein flexibility. 5) The creation of a more effective scoring function for HDAC with predictive power is also necessary for the rapid virtual screening of libraries of novel HDAC inhibitors.
ACKNOWLEDGEMENT
This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, and Center for Cancer Research. I thank Prof. Olaf Wiest for the review invitation and his encouragement and helps during this manuscript preparation. I am also very grateful to Mr. George Leiman, the manuscript reviewer and Dr. Victor Zhurkin for their critical reading, text corrections and valuable suggestions.
REFERENCES
- 1.(a) Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]; (b) Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]; (c) Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]; (d) Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]; (e) Yang WM, Inouye C, Zeng Y, Bearss D, Seto E. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator Rpd3. Proc. Natl. Acad. Sci. USA. 1996;93:1285–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Rundlett SE, Carmen AA, Kobayashi R, Bavykin S, Turner BM, Grunstein M. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl. Acad. Sci. USA. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu J, Grunstein M. 25 years after the nucleosome model: chromatin modifications. Trends Biochem. Sci. 2000;25:619–623. doi: 10.1016/s0968-0004(00)01718-7. [DOI] [PubMed] [Google Scholar]
- 3.Selected articles for HDAC related cancer diseases. Bulter LM, Agus DB, Scher HI, Higgins B, Rose A, Cordon-Cardo C, Thaler HT, Rifkind RA, Marks PA, Richon VM. Suberoylanilide hydroxamic acid, an inhibitor of histone deacetylase, suppresses the growth of prostate cancer cells in vitro and in vivo. Cancer Res. 2000;60:5166–5170.Halkidou K, Gaughan L, Cook S, Leung HY, Neal DE, Robson CN. Upregulation and nuclear recruitment of HDAC1 in hormone refractory prostate cancer. Prostate. 2004;59:177–189. doi: 10.1002/pros.20022.Wilson AJ, Byun DS, Popova N, Murray LB, L'Italien K, Sowa Y, Arango D, Velcich A, Augenlicht LH, Mariadason JM. Histone deacetylase 3 (HDAC3) and other class I HDACs regulate colon cell maturation and p21 expression and are deregulated in human colon cancer. J. Biol. Chem. 2006;281:13548–13558. doi: 10.1074/jbc.M510023200.Zhang Z, Yamashita H, Toyama T, Sugiura H, Ando Y, Mita K, Hamaguchi M, Hara Y, Kobayashi S, Iwase H. Breast Cancer Res. Treat. 2005;94:11–16. doi: 10.1007/s10549-005-6001-1.Zhang Z, Yamashita H, Toyama T, Sugiura H, Omoto Y, Ando Y, Mita K, Hamaguchi M, Hayashi S, Iwase H. HDAC6 expression is correlated with better survival in breast cancer. Clin. Cancer Res. 2004;10:6962–6968. doi: 10.1158/1078-0432.CCR-04-0455.Zhu P, Martin E, Mengwasser J, Schlag P, Janssen KP, Gottlicher M. Induction of HDAC2 expression upon loss of APC in colorectal tumorigenesis. Cancer Cell. 2004;5:455–463. doi: 10.1016/s1535-6108(04)00114-x.
- 4.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat. Rev. Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 5.Rasheed W, Bishton M, Johnstone RW, Prince HM. Histone deacetylase inhibitors in lymphoma and solid malignancies. Expert Rev. Anticancer Ther. 2008;8:413–432. doi: 10.1586/14737140.8.3.413. [DOI] [PubMed] [Google Scholar]; [b] Rasheed W, Johnstone RW, Prince HM. Histone deacetylase inhibitors in cancer therapy. Expert Opin. Investig. Drugs. 2007;16:659–678. doi: 10.1517/13543784.16.5.659. [DOI] [PubMed] [Google Scholar]
- 6.Kazantsev AG, Thompson LM. Nat. Rev. Drug Discov. 2008;7:854–868. doi: 10.1038/nrd2681. [DOI] [PubMed] [Google Scholar]
- 7.Meinke PT, Colletti SL, Doss G, Myers RW, Gurnett AM, Dulski PM, Darkin-Rattray SJ, Allocco JJ, Galuska S, Schmatz DM, Wyvratt MJ, Fisher MH. Synthesis of apicidin-derived quinolone derivatives: parasite-selective histone deacetylase inhibitors and antiproliferative agents. J. Med. Chem. 2000;43:4919–4922. doi: 10.1021/jm0001976. [DOI] [PubMed] [Google Scholar]
- 8.(a) De Ruijter AJ, Van Gennip AH, Caron HN, Kemp S, Van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Gregoretti I, Lee Y-M, Goodson HV. Molecular Evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J. Mol. Biol. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Ahringer J. NuRD and Sin3 histone deacetylase complexes in development. Trends Genet. 2000;16:351–356. doi: 10.1016/s0168-9525(00)02066-7. [DOI] [PubMed] [Google Scholar]
- 10.Wen YD, Perissi V, Staszewski LM, Yang W, Krones A, Glass CK, Rosenfeld MG, Seto E. The histone deacetylase-3 complex contains nuclear receptor corepressors. Proc. Natl. Acad. Sci. USA. 2000;97:7202–7207. doi: 10.1073/pnas.97.13.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.(a) Martin M, Kettmann R, Dequiedt F. Class IIa histone deacetylases: regulating the regulators. Oncogene. 2007;26:5450–5467. doi: 10.1038/sj.onc.1210613. [DOI] [PubMed] [Google Scholar]; (b) Majdzadeh N, Morrison BE, D'Mello SR. Class IIa HDACs in the regulation of neurodegeneration. Front. Biosci. 2008;13:1072–1082. doi: 10.2741/2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.(a) Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]; (b) Matsuyama A, Shimazu T, Sumida Y, Saito A, Yoshimatsu Y, Seigneurin-Berny D, Osada H, Komatsu Y, Nishino N, Khochbin S, Horinouchi S, Yoshida M. In vivo destabilization of dynamic microtubules by HDAC6-mediated deacetylation. EMBO J. 2002;21:6820–6831. doi: 10.1093/emboj/cdf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gray SG, Ekstrom TJ. The human histone deacetylase family. Exp. Cell Res. 2001;262:75–83. doi: 10.1006/excr.2000.5080. [DOI] [PubMed] [Google Scholar]
- 14.(a) Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem. J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Asher G, Gatfield D, Stratman M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]; (c) Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carew JS, Giles FJ, Nawrocki ST. Histone deacetylase inhibitors: mechanisms of cell death and promise in combination cancer therapy. Cancer Lett. 2008;269:7–17. doi: 10.1016/j.canlet.2008.03.037. [DOI] [PubMed] [Google Scholar]
- 16.Khan O, La Thangue NB. Drug insight: histone deacetylase inhibitor-based therapies for cutaneous T-cell lymphomas. Nat. Clin. Pract. Oncol. 2008 Oct 7; doi: 10.1038/ncponc1238. [DOI] [PubMed] [Google Scholar]
- 17.Butler KV, Kozikowski AP. Chemicla origins of isoform selectivity in histone deacetylase inhibitors. Curr. Pharm. Des. 2008;14:505–528. doi: 10.2174/138161208783885353. [DOI] [PubMed] [Google Scholar]
- 18.Finnin MS, Donigian JR, Cohen A, Richon VM, Rifkind RA, Marks PA, Breslow R, Pavletich NP. Structures of a Histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature. 1999;401:188–193. doi: 10.1038/43710. [DOI] [PubMed] [Google Scholar]
- 19.(a) Somoza JR, Skene RJ, Katz BA, Mol C, Ho JD, Jennings AJ, Luong C, Arvai A, Buggy JJ, Chi E, Tang J, Sang B-C, Verner E, Wynands R, Leahy EM, Dougan DR, Snell G, Navre M, Knuth MW, Swanson RV, Mcree DE, Tari LW. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure. 2004;12:1325–1334. doi: 10.1016/j.str.2004.04.012. [DOI] [PubMed] [Google Scholar]; (b) Vannini A, Volpari C, Filocamo G, Caroli Casavola E, Brunetti M, Renzoni D, Chakravarty P, Paolini C, De Francesco R, Gallinari P, Steinckuhler C, Di Marco S. Crystal structure of a eukaryotic Zn-Dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proc. Nat. Acad. Sci. USA. 2004;101:15064–15069. doi: 10.1073/pnas.0404603101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang D-F, Wiest O, Helquist P, Lan-Hargest HY, Wiech NL. On the function of the 14 Å long internal cavity of histone deacetylase-like protein: implications for the design of histone deacetylase inhibitors. J. Med. Chem. 2004;47:3409–3417. doi: 10.1021/jm0498497. [DOI] [PubMed] [Google Scholar]
- 21.Vannini A, Volpari C, Gallinari P, Jones P, Mattu M, Carfi A, De Francesco R, Steinkuhler C, Di Marco S. Substrate binding to histone deacetylases as shown by the crystal structure of the HDAC8–substrate complex. EMBO Rep. 2007;8:879–884. doi: 10.1038/sj.embor.7401047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schuetz A, Min J, Allali-Hassani A, Schapira M, Shuen M, Loppnau P, Mazitschek R, Kwiatkowski NP, Lewis TA, Maglathin RL, McLean TH, Bochkarev A, Plotnikov AN, Vedadi M, Arrowsmith CH. Human HDAC7 harbors a class IIa histone deacetylase-specific Zinc binding motif and cryptic deacetylase activity. J. Biol. Chem. 2008;283:11355–11363. doi: 10.1074/jbc.M707362200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bottomley MJ, Lo Surdo P, Di Giovine P, Cirillo A, Scarpelli R, Federica Ferrigno F, Jones P, Neddermann P, De Francesco R, Steinkuhler C, Gallinari P, Carfi A. Structural and functional analysis of the human HDAC4 catalytic domain reveals a regulatory structural Zinc-binding domain. J. Biol. Chem. 2008;283:26694–26704. doi: 10.1074/jbc.M803514200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nielsen TK, Hildmann C, Dickmanns A, Schwienhorst A, Ficner R. Crystal structure of a bacterial class 2 histone deacetylase homologue. J. Mol. Biol. 2005;354:107–120. doi: 10.1016/j.jmb.2005.09.065. [DOI] [PubMed] [Google Scholar]
- 25.Finnin MS, Donigian JR, Pavletich NP. Structure of the histone deacetylase SIRT2. Nat. Struct. Biol. 2001;8:621–625. doi: 10.1038/89668. [DOI] [PubMed] [Google Scholar]
- 26.Schuetz A, Min J, Antoshenko T, Wang CL, Allali-Hassani A, Dong A, Loppnau P, Vedadi M, Bochkarev A, Sternglanz R, Plotnikov AN. Structural basis of inhibition of the human NAD+-dependent deacetylase SIRT5 by suramin. Structure. 2007;15:377–389. doi: 10.1016/j.str.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 27.Zhao K, Chai X, Marmorstein R. Structure of the yeast Hst2 protein deacetylase in ternary complex with 2′-O-acetyl ADP ribose and histone peptide. Structure. 2003;11:1403–1411. doi: 10.1016/j.str.2003.09.016. [DOI] [PubMed] [Google Scholar]
- 28.Avalos JL, Bever KM, Wolberger C. Structural basis for the mechanism and regulation of Sir2 enzymes. Mol. Cell. 2005;17:855–868. doi: 10.1016/s1097-2765(04)00082-6. [DOI] [PubMed] [Google Scholar]
- 29.Avalos JL, Boeke JD, Wolberger C. Mechanism of sirtuin inhibition by nicotinamide: altering the NAD(+) cosubstrate specificity of a Sir2 enzyme. Mol. Cell. 2004;13:639–648. doi: 10.1016/j.molcel.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 30.Chang JH, Kim HC, Hwang KY, Lee JW, Jackson SP, Bell SD, Cho Y. Structural basis for the NAD-dependent deacetylase mechanism of Sir2. J. Biol. Chem. 2002;277:34489–34498. doi: 10.1074/jbc.M205460200. [DOI] [PubMed] [Google Scholar]
- 31.Zhao K, Chai X, Marmorstein R. Structure and substrate binding properties of cobB, a Sir2 homolog protein deacetylase from Escherichia coli. J. Mol. Biol. 2004;337:731–741. doi: 10.1016/j.jmb.2004.01.060. [DOI] [PubMed] [Google Scholar]
- 32.Park H, Lee SJ. Homology modeling, force field design, and free energy simulation studies to optimize the activities of histone deacetylase inhibitors. J. Comput. Aided Mol. Des. 2004;18:375–388. doi: 10.1007/s10822-004-2283-3. [DOI] [PubMed] [Google Scholar]
- 33.Massa S, Mai A, Sbardella G, Esposito M, Ragno R, Loidl P, Brosch G. 3-(4-aroyl-1H-pyrrol-2-yl)-N-hydroxy-2-propenamides, a new class of synthetic histone deacetylase inhibitors. J. Med. Chem. 2001;44:2069–2072. doi: 10.1021/jm015515v. [DOI] [PubMed] [Google Scholar]
- 34.(a) Mai A, Massa S, Ragno R, Cerbara I, Jesacher F, Loidl P, Brosch G. 3-(4-Aroyl-1-methyl-1H-2-pyrrolyl)-N-hydroxy-2-alkylamides as a new class of synthetic histone deacetylase inhibitors. 1. Design, synthesis, biological evaluation, and binding mode studies performed through three different docking procedures. J. Med. Chem. 2003;46:512–524. doi: 10.1021/jm021070e. [DOI] [PubMed] [Google Scholar]; (b) Mai A, Massa S, Cerbara I, Valente S, Ragno R, Bottoni P, Scatena R, Loidl P, Brosch G. 3-(4-Aroyl-1-methyl-1H-2-pyrrolyl)-N-hydroxy-2-propenamides as a new class of synthetic histone deacetylase inhibitors. 2. Effect of pyrrole-C2 and/or -C4 substitutions on biological activity. J. Med. Chem. 2004;47:1098–1109. doi: 10.1021/jm030990+. [DOI] [PubMed] [Google Scholar]; (c) Ragno R, Mai A, Massa S, Cerbara I, Valente S, Bottoni P, Scatena R, Jesacher F, Loidl P, Brosch G. 3-(4-Aroyl-1-methyl-1H-pyrrol-2-yl)-N-hydroxy-2-propenamides as a new class of synthetic histone deacetylase inhibitors. 3. Discovery of novel lead compounds through structure-based drug design and docking studies. J. Med. Chem. 2004;47:1351–1359. doi: 10.1021/jm031036f. [DOI] [PubMed] [Google Scholar]
- 35.Vanommeslaeghe K, Loverix S, Geerlings P, Tourwe D. DFT-based ranking of zinc-binding groups in histone deacetylase inhibitors. Bioorg. Med. Chem. 2005;13:6070–6082. doi: 10.1016/j.bmc.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 36.Wang D-F, Helquist P, Wiest O. Zinc binding in HDAC inhibitors: A DFT study. J. Org. Chem. 2007;72:5446–5449. doi: 10.1021/jo070739s. [DOI] [PubMed] [Google Scholar]
- 37.Wang D-F, Helquist P, Wiech NL, Wiest O. Toward selective histone deacetylase inhibitor design: homology modeling, docking studies, and molecular dynamics simulations of human class I histone deacetylases. J. Med. Chem. 2005;48:6936–47. doi: 10.1021/jm0505011. [DOI] [PubMed] [Google Scholar]
- 38.(a) Moradei OM, Mallais TC, Frechette S, Paquin I, Tessier PE, Leit SM, Fournel M, Bonfils C, Trachy-Bourget MC, Liu J, Yan TP, Lu AH, Rahil J, Wang J, Lefebvre S, Li Z, Vaisburg AF, Besterman JM. Novel aminophenyl benzamide-type histone deacetylase inhibitors with enhanced potency and selectivity. J. Med. Chem. 2007;50:5543–5546. doi: 10.1021/jm701079h. [DOI] [PubMed] [Google Scholar]; (b) Methot JL, Chakravarty PK, Chenard M, Close J, Cruz JC, Dahlberg WK, Fleming J, Hamblett CL, Hamill JE, Harrington P, Harsch A, Heidebrecht R, Hughes B, Jung J, Kenific CM, Kral AM, Meinke PT, Middleton RE, Ozerova N, Sloman DL, Stanton MG, Szewczak AA, Tyagarajan S, Witter DJ, Secrist JP, Miller TA. Exploration of the internal cavity of histone deacetylase (HDAC) with selective HDAC1/HDAC2 inhibitors (SHI-1:2) Bioorg. Med. Chem. Lett. 2008;18:973–978. doi: 10.1016/j.bmcl.2007.12.031. [DOI] [PubMed] [Google Scholar]
- 39.Kim HM, Hong SH, Kim MS, Lee CW, Kang JS, Lee K, Park SY, Han JW, Lee HY, Choi Y, Kwon HJ, Han G. Modification of cap group in δ-lactam-based histone deacetylase (HDAC) inhibitors. Bioorg. Med. Chem. Lett. 2007;17:6234–6238. doi: 10.1016/j.bmcl.2007.09.034. [DOI] [PubMed] [Google Scholar]
- 40.Hamblett CL, Methot JL, Mampreian DM, Sloman DL, Stanton MG, Kral AM, Fleming JC, Cruz JC, Chenard M, Ozerova N, Hitz AM, Wang H, Deshmukh SV, Nazef N, Harsch A, Hughes B, Dahlberg WK, Szewczak AA, Middleton RE, Mosley RT, Secrist JP, Miller TA. The discovery of 6-amino nicotinamides as potent and selective histone deacetylase inhibitors. Bioorg. Med. Chem. Lett. 2007;17:5300–5309. doi: 10.1016/j.bmcl.2007.08.023. [DOI] [PubMed] [Google Scholar]
- 41.Nakao Y, Yoshida S, Matsunaga S, Shindoh N, Terada Y, Nagai K, Yamashita JK, Ganesan A, van Soest RWM, Fusetani N. Azumamides A-E: histone deacetylase inhibitory cyclic tetrapeptides from the marine sponge Mycale izuensis. Angew. Chem. Int. Ed. 2006;45:7553–7557. doi: 10.1002/anie.200602047. [DOI] [PubMed] [Google Scholar]; b) Izzo I, Maulucci N, Bifulco G, De Riccardis F. Total synthesis of azumamides A and E. Angew. Chem. Int. Ed. 2006;45:7557–7560. doi: 10.1002/anie.200602033. [DOI] [PubMed] [Google Scholar]
- 42.Maulucci N, Chini MG, Micco SD, Izzo I, Cafaro E, Russo A, Gallinari P, Paolini C, Nardi MC, Casapullo A, Riccio R, Bifulco G, Riccardis FD. Molecular insights into azumamide E histone deacetylases inhibitory activity. J. Am. Chem. Soc. 2007;129:3007–3012. doi: 10.1021/ja0686256. [DOI] [PubMed] [Google Scholar]
- 43.Weerasinghe SV, Estiu G, Wiest O, Pflum MK. Residues in the 11 Å channel of histone deacetylase 1 promote catalytic activity: implications for designing isoform-selective histone deacetylase inhibitors. J. Med. Chem. 2008;51:5542–5551. doi: 10.1021/jm800081j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Itoh Y, Suzuki T, Miyata N. Isoform-selective histone deacetylase inhibitors. Curr. Pharm. Des. 2008;14:529–544. doi: 10.2174/138161208783885335. [DOI] [PubMed] [Google Scholar]
- 45.Butler KV, Kozikowski AP. Chemical origins of isoform selectivity in histone deacetylase inhibitors. Curr. Pharm. Des. 2008;14:505–528. doi: 10.2174/138161208783885353. [DOI] [PubMed] [Google Scholar]
- 46.Haggarty SJ, Koeller KM, Wong JC, Grozinger CM, Schreiber SL. Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc. Natl. Acad. Sci. USA. 2003;100:4389–4394. doi: 10.1073/pnas.0430973100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suzuki T, Kouketsu A, Itoh Y, Hisakawa S, Maeda S, Yoshida M, Nakagawa H, Miyata N. Highly potent and selective histone deacetylase 6 inhibitors designed based on a small-molecular substrate. J. Med. Chem. 2006;49:4809–4812. doi: 10.1021/jm060554y. [DOI] [PubMed] [Google Scholar]
- 48.Mai A, Massa S, Pezzi R, Simeoni S, Rotili D, Nebbioso A, Scognamiglio A, Altucci L, Loidl P, Brosch G. Class II (IIa)-selective histone deacetylase inhibitors. 1. Synthesis and biological evaluation of novel (aryloxopropenyl)pyrrolyl hydroxyamides. J. Med. Chem. 2005;48:3344–3353. doi: 10.1021/jm049002a. [DOI] [PubMed] [Google Scholar]
- 49.Schafer S, Saunders L, Eliseeva E, Velena A, Jung M, Schwienhorst A, Strasser A, Dickmanns A, Ficner R, Schlimme S, Sippl W, Verdin E, Jung M. Phenylalanine-containing hydroxamic acids as selective inhibitors of class IIb histone deacetylases (HDACs) Bioorg. Med. Chem. 2008;16:2011–2033. doi: 10.1016/j.bmc.2007.10.092. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Y, Gilquin B, Khochbin S, Matthias P. Two catalytic domains are required for protein deacetylation. J. Biol. Chem. 2006;281:2401–2404. doi: 10.1074/jbc.C500241200. [DOI] [PubMed] [Google Scholar]
- 51.Zou H, Wu Y, Navre M, Sang BC. Characterization of the two catalytic domains in histone deacetylase 6. Biochem. Biophys. Res. Commun. 2006;341:45–50. doi: 10.1016/j.bbrc.2005.12.144. [DOI] [PubMed] [Google Scholar]
- 52.Kozikowski AP, Tapadar S, Luchini DN, Kim KH, Billadeau DD. Use of the nitrile oxide cycloaddition (NOC) reaction for molecular probe generation: a new class of enzyme selective histone deacetylase inhibitors (HDACIs) showing picomolar activity at HDAC6. J. Med. Chem. 2008;51:4370–4373. doi: 10.1021/jm8002894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Estiu G, Greenberg E, Harrison CB, Kwiatkowski NP, Mazitschek R, Bradner JE, Wiest O. Structural origin of selectivity in class II-selective histone deacetylase inhibitors. J. Med. Chem. 2008;51:2898–2906. doi: 10.1021/jm7015254. [DOI] [PubMed] [Google Scholar]
- 54.Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 55.Porcu M, Chiarugi A. The emerging therapeutic potential of sirtuin-interacting drugs: from cell death to lifespan extension. Trends Pharmacol. Sci. 2005;26:94–103. doi: 10.1016/j.tips.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 56.Neugebauer RC, Sippl W, Jung M. Inhibitors of NAD+ dependent histone deacetylases (sirtuins) Curr. Pharm. Des. 2008;14:562–573. doi: 10.2174/138161208783885380. [DOI] [PubMed] [Google Scholar]
- 57.Huhtiniemi T, Wittekindt C, Laitinen T, Leppanen J, Salminen A, Poso A, Lahtela-Kakkonen M. Comparative and pharmacophore model for deacetylase SIRT1. J. Comput. Aided Mol. Des. 2006;20:589–599. doi: 10.1007/s10822-006-9084-9. [DOI] [PubMed] [Google Scholar]
- 58.Neugebauer RC, Uchiechowska U, Meier R, Hruby H, Valkov V, Verdin E, Sippl W, Jung M. Structure-activity studies on splitomicin derivatives as sirtuin inhibitors and computational prediction of binding mode. J. Med. Chem. 2008;51:1203–1213. doi: 10.1021/jm700972e. [DOI] [PubMed] [Google Scholar]
- 59.Heltweg B, Gatbonton T, Schuler AD, Posakony J, Li HZ, Goehle S, Kollipara R, Depinho RA, Gu Y, Simon JA, Bedalov A. Antitumor activity of a small-molecule inhibitor of human silent information regulator 2 enzymes. Cancer Res. 2006;66:4368–4377. doi: 10.1158/0008-5472.CAN-05-3617. [DOI] [PubMed] [Google Scholar]
- 60.Qikprop 1.6, Schrodinger Inc. 2001. to calculate the properties. For publications, see Duffy EM, Jorgensen WL. Prediction of properties from simulations: free energies of solvation in hexadecane, octanol, and water. J. Am. Chem. Soc. 2000;122:2878–2888.Jorgensen WL, Duffy EM. Prediction of drug solubility from Monte Carlo simulations. Bioorg. Med. Chem. Lett. 2000;10:1155–1158. doi: 10.1016/s0960-894x(00)00172-4.
- 61.Wang D-F, Wiest O, Helquist P, Lan-Hargest HY, Wiech NL. QSAR studies of PC-3 cell line inhibition activity of TSA and SAHA-like hydroxamic acids. Bioorg. Med. Chem. Lett. 2004;14:707–711. doi: 10.1016/j.bmcl.2003.11.062. [DOI] [PubMed] [Google Scholar]
- 62.Xie A, Liao C, Li Z, Ning Z, Hu W, Lu X, Shi L, Zhou J. Quantitative structure-activity relationship study of histone deacetylase inhibitors. Curr. Med. Chem. Anticancer Agents. 2004;4:273–299. doi: 10.2174/1568011043352948. [DOI] [PubMed] [Google Scholar]
- 63.Kozikowski AP, Chen Y, Gaysin AM, Savoy DN, Billadeau DD, Kim KH. Chemistry, biology, and QSAR studies of substituted biaryl hydroxamates and mercaptoacetamides as HDAC inhibitors-nanomolar-potency inhibitors of pancreatic cancer cell growth. ChemMedChem. 2008;3:487–501. doi: 10.1002/cmdc.200700314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen H-F, Kang J-H, Li Q, Zeng B-S, Yao X-J, Fan B-T, Yuan S-G, Panay A, Doucet JP. 3D-QSAR study on apicidin inhibit histone deacetylase. Chin. J. Chem. 2003;21:1596–1607. [Google Scholar]
- 65.Guo Y, Xiao J, Guo Z, Chu F, Cheng Y, Wu S. Exploration of a binding mode of indole amide analogues as potent histone deacetylase inhibitors and 3D-QSAR analyses. Bioorg. Med. Chem. 2005;13:5424–5434. doi: 10.1016/j.bmc.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 66.Juvale DC, Kulkarni VV, Deokar HS, Wagh NK, Padhye SB, Kulkarni VM. 3D-QSAR of histone deacetylase inhibitors: hydroxamate analogues. Org. Biomol. Chem. 2006;15:2858–2868. doi: 10.1039/b606365a. [DOI] [PubMed] [Google Scholar]
- 67.Chen YD, Jiang YJ, Zhou JW, Yu QS, You QD. Identification of ligand features essential for HDACs inhibitors by pharmacophore modeling. J. Mol. Graph. Model. 2008;26:1160–1168. doi: 10.1016/j.jmgm.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 68.Ragno R, Simeoni S, Rotili D, Caroli A, Botta G, Brosch G, Massa S, Mai A. Class II-selective histone deacetylase inhibitors. Part 2: Alignment-independent GRIND 3-D QSAR, homology and docking studies. Eur. J. Med. Chem. 2008;43:621–632. doi: 10.1016/j.ejmech.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 69.Avalos JL, Celic I, Muhammad S, Cosgrove MS, Boeke JD, Wolberger C. Structure of a Sir2 enzyme bound to an acetylated p53 peptide. Mol. Cell. 2002;10:523–535. doi: 10.1016/s1097-2765(02)00628-7. [DOI] [PubMed] [Google Scholar]
- 70.Min J, Landry J, Sternglanz R, Xu RM. Crystal structure of a SIR2 homolog-NAD complex. Cell. 2001;105:269–276. doi: 10.1016/s0092-8674(01)00317-8. [DOI] [PubMed] [Google Scholar]