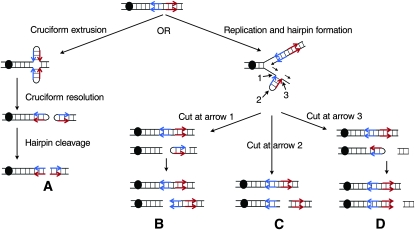
Figure 1.—
Mechanisms of producing a recombinogenic DSB at an IR. The inverted repeat is shown as blue and red arrows, and each line represents a single DNA strand rather than duplicated chromatids. Labeled arrows show the positions of nuclease cleavage at the hairpin structure. The centromere is shown as a black oval. Only those broken DNA molecules containing a centromere are likely to produce a recoverable chromosome rearrangement. (A) Cruciform formation in a nonreplicating DNA molecule. Processing of the resulting structure by a resolvase would be expected to yield two hairpin-capped products that could be subsequently processed to yield uncapped broken DNA molecules. (B–D) DSBs produced by different positions of cleavage of the hairpin intermediate. We show hairpin formation associated with replication of the lagging strand. Cleavage at arrow 1 produces a capped hairpin in the acentric fragment or a centromere-containing fragment with a DSB proximal to FS2. Cleavage at arrow 2 results in a product in which the DSB is between the two elements of the inverted repeat. Cleavage at arrow 3 produces a capped hairpin or, if replication proceeds through the hairpin, results in a centromere-containing fragment with a DSB near the distal Ty element of FS2.