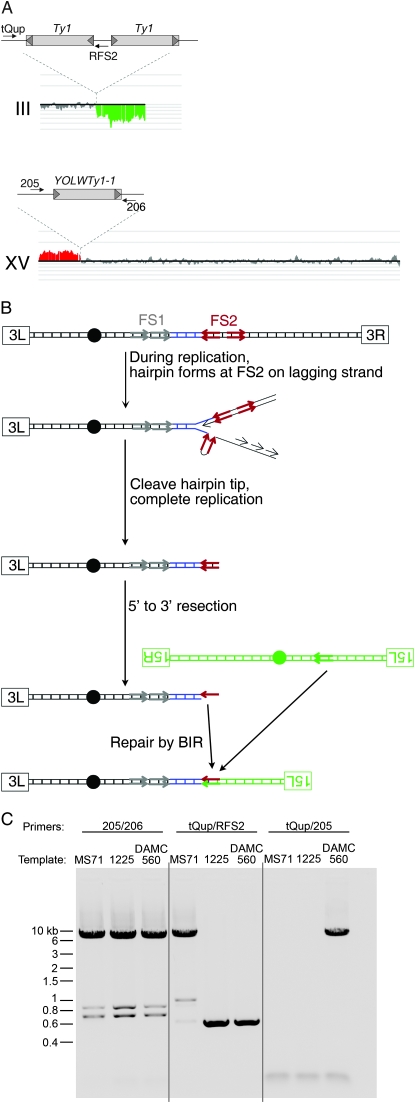
Figure 5.—
Physical analysis of a translocation produced by a BIR event between nonallelic Ty elements in an mre11Δ/ mre11Δ illegitimate diploid. (A) Microarray analysis. DNA was isolated from a class 3 illegitimate diploid (DAMC560) resulting from the mating of two MATα mre11Δ strains. This sample was labeled with a Cy5 fluorescent nucleotide and mixed with a control DNA sample labeled with a Cy3 fluorescent nucleotide, and this mixture was used a hybridization probe of a microarray containing all of the yeast ORFs and intergenic regions. The ratios of hybridization are indicated as vertical lines with deletions and additions in the experimental strain shown in green and red, respectively (analysis by the CGH-Miner program). No changes were observed on chromosomes other than III and XV. The deletion breakpoint on III is at FS2, and the amplification breakpoint on XV is at YOLWTy1-1. Large gray rectangles represent Ty elements, short gray arrowheads show δ-elements, and small black arrows represent PCR primers. (B) Mechanism for generating the III–XV translocation by BIR. Centromeres are indicated by black circles, left and right telomeres are identified by labeled rectangles, and Ty elements are indicated by arrows. (C) Confirmation of translocation by PCR. The positions of the primers are shown in A. MS71 is the wild-type parental haploid from which all GAL-POL1 experimental strains are derived, and 1225 is the wild-type parental haploid from which all mating-type tester strains are derived. MS71 has the centromere-distal Ty at FS2 that 1225 lacks. As expected, PCR using primers from III (tQup) and XV (205) generates a product when DNA from the strain with the III–XV translocation is used as a template.