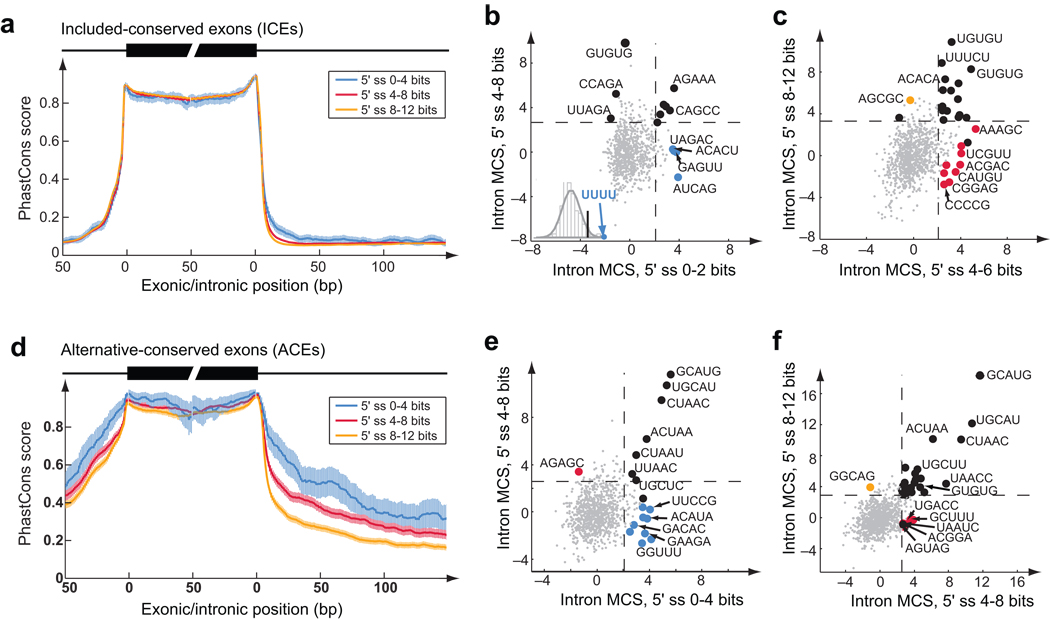
Figure 4.
Sequence conservation flanking exons dependent on 5’ss strength. (a) Conservation profile (mean and 95% confidence interval of PhastCons scores49) of exons and flanking introns for orthologous human/mouse included-conserved exons (ICEs) grouped by 5’ss score (bits). (b) Motif conservation scores (MCS, Supplementary Methods) of 5mers in downstream introns (11–70 nt from 5’ss) of ICEs with indicated 5’ss scores. Dashed lines show cutoff of significant MCS determined based on randomly shuffled data. Black dots represent motifs that are significantly conserved in more than one 5’ss groups. Motifs with more significant MCS in one group than all other groups are represented by colored dots in the same color scheme as in (a). Inset shows the histogram of the t-statistic of 4mers between the two indicated 5’ss groups. UUUU was the most significant in the 0–2 bits group. (c) Same as (b) for the 5’ss groups of 4–6 and 8–12 bits. (d, e, f) same as (a, b, c) for alternative-conserved exons (ACEs). Scatter plots are shown for the downstream intronic region 11–200 nt from the 5’ss. Because the number of exons was smaller in this analysis (∼3,000), only 3 bins of 5’ss strength were used (0–4, 4–8, 8–12 bits).