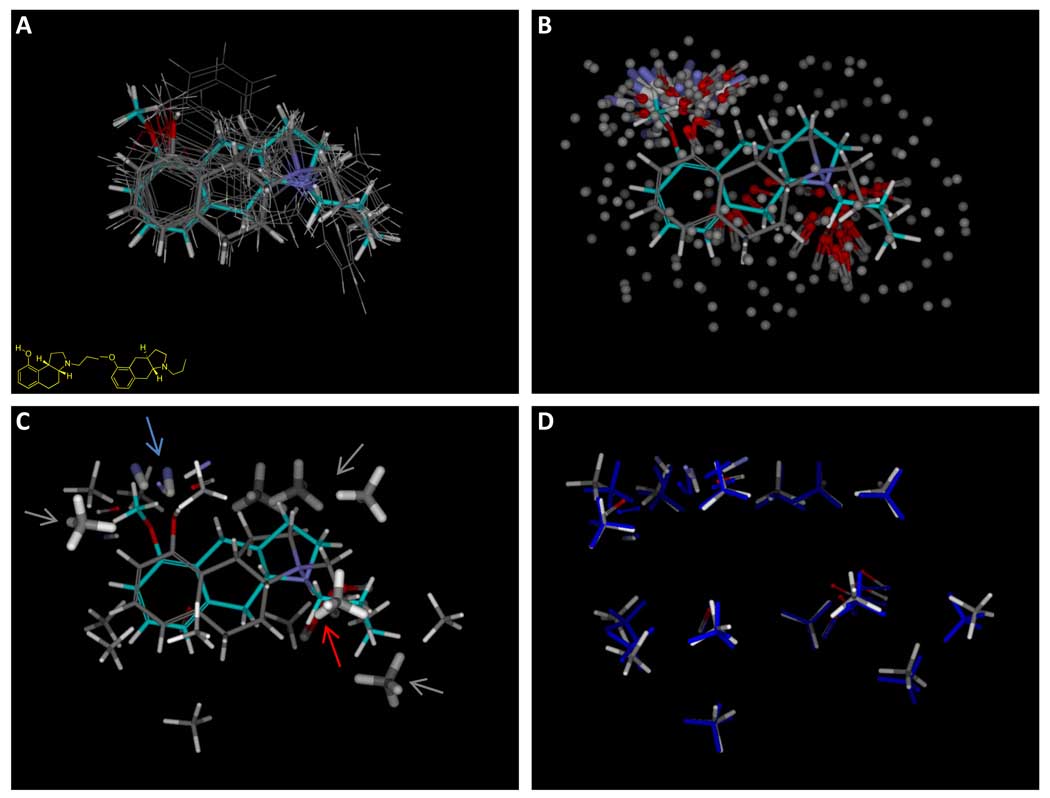
Figure 7.
The overall procedure begins with an initial alignment hypothesis, as shown in Figure 2, which is used to generate initial alignments of all training ligands, possibly using multiple approaches (panel A shows a single alignment for each active training ligand). The initial alignments of active ligands are used to produce a large number of molecular probes that interact well with at least one pose of one active ligand (panel B, with hydrophobic probes shown without hydrogens for clarity). A subset of the probes are chosen to optimize concordance with activity (panel C, thick sticks). These identify the known pharmacophore, characterized by interactions with a donor (blue arrow), a charged acceptor (red arrow), and hydrophobic interactions (gray arrows). Additional probes are added back in order to provide “coverage” of the entire possible pocket (probes shown with thin sticks). Panel D shows the final optimized pocket (atom color) along with the initial probe positions (blue).