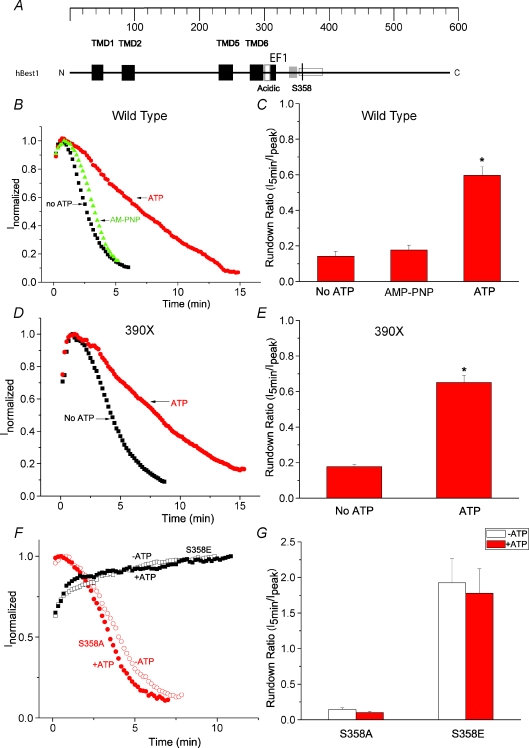
Figure 1. Identification of S358 as a phosphorylation site in hBest1.
A, model of functional domains in hBest1. Black (TMD): transmembrane domains (Milenkovic et al. 2007). Open (acidic): acidic domain. Black (EF1): EF1 domain, critical for Ca2+ binding. Open: a region identified to be responsible for channel rundown (amino acids 350–390) (Xiao et al. 2008). S358 is indicated by a black line. An SH3-binding domain between EF1 and S358 was identified by Yu et al. 2008 (Yu et al. 2008). B, D and F, time course of typical currents of wild-type (B), 390X mutant (D) and S358E and S358A mutants (F). Voltage ramps from −100 mV to +100 mV were given from a holding potential of 0 mV at 10 s intervals. The currents at +100 mV (filled symbols) were normalized to the peak current. B, wild-type hBest1 currents recorded in the presence of ATP (circles), no ATP (squares), and AMP-PNP (triangles). D, 390X mutant currents recorded in the presence of ATP (circles), and no ATP (squares). The 390X mutant was made by introducing stop codon at amino acid 390. F, S358E mutant (squares) and S358A mutant (circles) currents recorded in the presence of ATP (filled symbols) and no ATP (open symbols). C, E and G, average values of effects of ATP on channel rundown of wild-type hBest1 (C, n= 8–12), 390X mutant (E, n= 5–6) and S358E and S358A mutants (G, n= 4–7). The rundown was measured by the ratio of amplitudes of the current 5 min after peak current to the amplitudes at the peak. *P < 0.05 versus no ATP by one-way ANOVA.