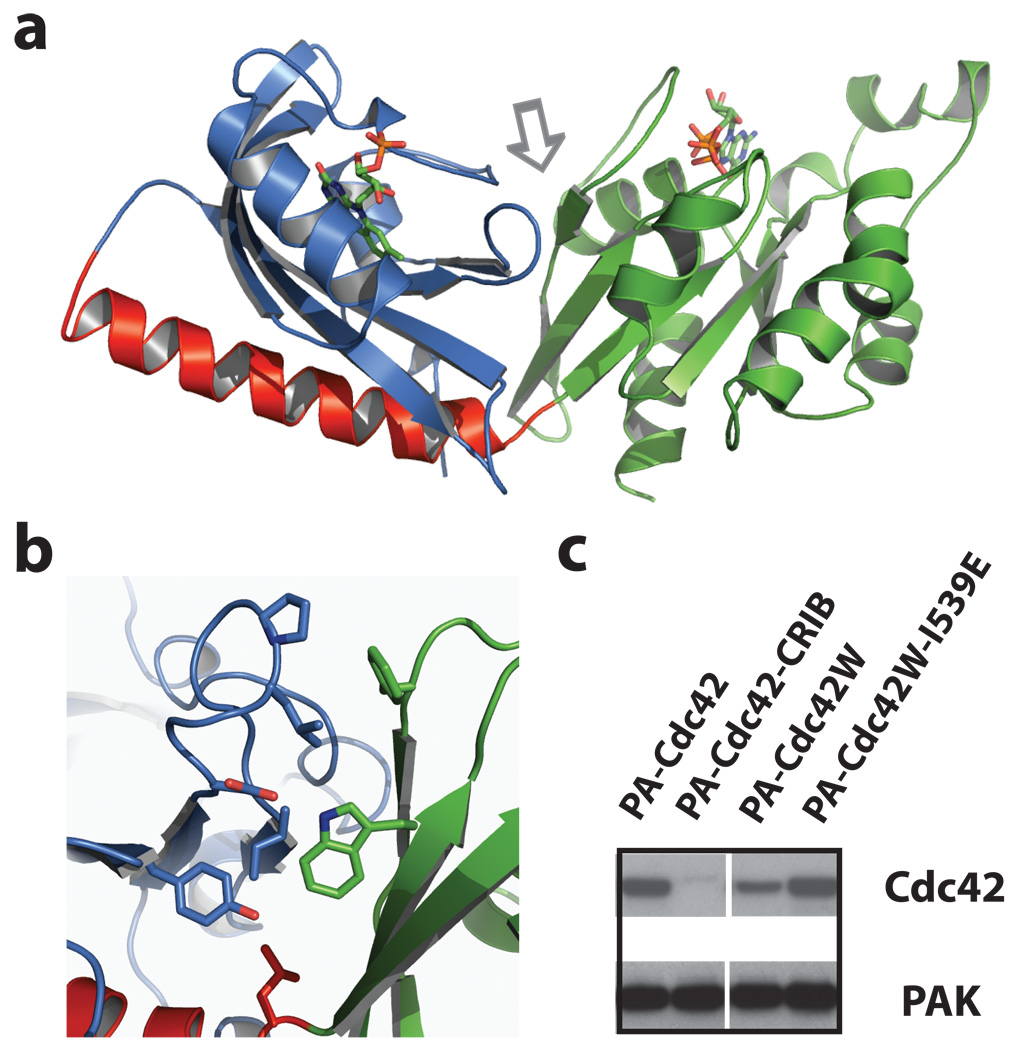
Figure 4. Crystallization and structural modelling of PA-Rac1.
a, Dark state crystal structure of PA-Rac1. Blue = LOV domain, red = Jα helix, and green = Rac1. b, Interacting residues at the LOV-Rac interface (arrow in panel a), including Trp56. c, Mutating Cdc42 to include the Trp involved in stabilizing the LOV2-Rac1 interaction substantially improved LOV inhibition of Cdc42. Lane 1, PA-Cdc42; linking LOV to Cdc42 using the same truncations that produced good inhibition for Rac does not inhibit Cdc42-PAK binding. Lane 2, PA-Cdc42-CRIB; covalently linking the CRIB domain of PAK to PA-Cdc42 blocks PAK binding. Lane 3, PA-Cdc42-F56W; introduction of the tryptophan substantially improves LOV inhibition of Cdc42 binding to PAK. Lane 4, lit state mutant of PA-Cdc42-F56W, showing that Cdc42 inhibition is sensitive to the lit/dark state of the LOV domain. Supplemental Movie S16 and Fig. S14 demonstrate the ability of PA-Cdc42-F56W to produce filopodia and protrusions in living cells.