Abstract
Chemotherapy for metastatic bladder cancer is rarely curative. The recently developed small molecule, lapatinib, a dual epidermal growth factor receptor (EGFR)/human epidermal growth factor receptor-2 receptor tyrosine kinase inhibitor, might improve this situation. Recent findings suggest that identifying which patients are likely to benefit from targeted therapies is beneficial, although controversy remains regarding what types of evaluation might yield optimal candidate biomarkers of sensitivity. Here, we address this issue by developing and comparing lapatinib sensitivity prediction models for human bladder cancer cells. After empirically determining in vitro sensitivities (drug concentration necessary to cause a 50% growth inhibition) of a panel of 39 such lines to lapatinib treatment, we developed prediction models based on profiling the baseline transcriptome, the phosphorylation status of EGFR pathway signaling targets, or a combination of both data sets. We observed that models derived from microarray gene expression data showed better prediction performance (93%–98% accuracy) compared with models derived from EGFR pathway profiling of 23 selected phosphoproteins known to be involved in EGFR-driven signaling (54%–61% accuracy) or from a subset of the microarray data for transcripts in the EGFR pathway (86% accuracy). Combining microarray data and phosphoprotein profiling provided a combination model with 98% accuracy. Our findings suggest that transcriptome-wide profiling for biomarkers of lapatinib sensitivity in cancer cells provides models with excellent predictive performance and may be effectively combined with EGFR pathway phosphoprotein profiling data. These results have significant implications for the use of such tools in personalizing the approach to cancers treated with EGFR-directed targeted therapies.
Introduction
Bladder cancer is the fourth most common tumor diagnosed in the United States [1] and among the most expensive cancers to treat [2]. Approximately 50% of bladder cancer patients that present with muscle invasive disease develop metastasis [3]. Systemic chemotherapy with the methotrexate, vinblastine, doxorubicin, and cisplatin regimen has been used for more than a decade in patients producing a median survival of 12 months [4]. Newer combinations such as gemcitabine-cisplatin are gaining popularity given their diminished toxicity, yet they do not offer improvements in clinical outcome [5]. Hence, there is a strong impetus to discover more effective agents for advanced bladder cancer whether to be used alone or in combination with established drugs in the metastatic setting.
Epidermal growth factor receptor (EGFR) has been associated with bladder tumor progression and hence is an important potential target for therapy for invasive or metastatic bladder cancer [6]. The EGFR family contains four distinct but structurally similar tyrosine kinase receptors encoded by the genes EGFR, ERBB2 (human epidermal growth factor receptor-2 [HER2]), ERBB3 (HER3), and ERBB4 (HER4). Generally, these receptors possess extracellular, transmembrane, and intracellular tyrosine kinase domains. After ligand binding and formation of homodimers or heterodimers among family members, the ligand-receptor complex undergoes tyrosine autophosphorylation in trans, resulting in signaling that supports cell survival, angiogenesis, and even metastatic activity of tumors [7]. Studies have reported associations between EGFR and HER2 expression with both higher bladder tumor grade and invasion [8], whereas expression of HER2 alone, or in association with EGFR and HER3, was negatively associated with patient survival [9]. Experimental models have also confirmed a role for these receptors in bladder cancer cell migration and other surrogates of metastasis [10,11].
Lapatinib (Tykerb, GW572016) is a reversible noncovalent inhibitor of both EGFR and HER2 receptors that has shown promise in clinical trials in breast and other cancers [12]. Expression of HER2 has been established as a minimal requirement for patient inclusion in previous breast cancer trials [13]. Beyond that, there remains significant disagreement regarding whether expression or phosphorylation level, genetic amplification, or other measures of HER2 or EGFR expression and activity may predict response to lapatinib [14]. In addition, a recent study of 61 human tumor cell lines from 12 different histologies found that expression levels of both EGFR andHER2 have to be considered for prediction, along with the type of tissue of origin, to provide the best prediction of response to lapatinib [15]. A phase 2 clinical trial of lapatinib in inflammatory breast carcinoma found that expression of phosphorylated HER3 and lack of p53 expression independently associated with response, whereas patients whose tumors coexpressed phosphorylated HER2 and HER3 were more likely to respond than tumors that did not [16]. Together, these findings suggest assessing the activity of EGFR family signaling pathways to predict lapatinib sensitivity.
Profiling cells using genomic technologies has also predicted drug sensitivity. Recently, we generated gene expression signatures of sensitivity to three common chemotherapeutic agents, namely, gemcitabine, cisplatin, and paclitaxel, finding that such signatures predict sensitivity in human bladder cancer cells [17]. Another group has developed, based on in vitro cell line testing, microarray-based signatures to predict in vitro sensitivity of cell lines to erlotinib and found that such signatures can predict EGFR activation and mutation in human lung cancer clinical samples [18]. Such findings suggest that predictive biomarkers of lapatinib sensitivity might also be discovered through high throughput gene expression analysis.
These findings raise important and timely questions. First, can gene expression profiling and EGFR pathway phosphoprotein profiling provide useful tools to predict response to lapatinib? Second, if both approaches provide some prediction, can a combination of both provide even better results? Answering these questions would lay the conceptual framework for a practical implementation of cancer evaluation for the prediction of EGFR-targeted therapeutics. Here, we use a panel of human bladder cancer cell lines as a model to answer these questions.
Materials and Methods
Cell Lines, Cell Culture, and Generation of Lapatinib Response Data
Human bladder cancer cell lines used in this study and their growth conditions were previously described [17,19–21]. Lapatinib was provided by GlaxoSmithKline, Research Triangle Park, NC, dissolved in 100% DMSO, and aliquoted as 8-mM stock until use. Cell lines were seeded at a density of 1000 cells per well in 96-well cell culture plates. After 24 hours, lapatinib was added to each well in RPMI-1640 plus 10% FBS (all cell lines used were tested in this standard medium for the sake of internal consistency), in a total volume of 200 µl. Lapatinib doses used were 0.25, 0.5, 1, 2, 4, and 8 µM. Every dose of lapatinib was tested in triplicate, and each experiment was repeated four to seven times for each cell line. Each 96-well plate contained a row of 8 wells with RPMI-1640 media plus 10% FBS and 0.1% DMSO as an untreated control. Plates were incubated for 72 hours with media only or the above lapatinib doses, after which growth inhibition was assessed by Alamar Blue (BioSource International, Camarillo, CA) [17,22]. For immunoprecipitation experiments, cells were exposed to either DMSO vehicle or lapatinib at their calculated drug concentration necessary to cause a 50% growth inhibition (GI50; see below) for 24 hours, conventional immunoprecipitations were carried out with anti-phosphotyrosine antibodies, then precipitates were immunoblotted for total EGFR (both antibodies from Cell Signaling Technologies, Danvers, MA).
Preparation of Cells for Phosphoproteomic Analysis
Bladder cancer cells were seeded in 100-mm cell culture plates. After reaching 80%to 90%confluence, cells were rinsed with 5 ml of ice-cold PBS. Cells were rapidly scraped using a rubber policeman and transferred to microcentrifuge tubes. Cell pellets were collected, snap-frozen with liquid nitrogen, and stored at -80°C until analysis.
Gene Expression and Reverse-Phase Protein Microarray Profiling of Bladder Cell Lines
Gene expression profiling data of the 39 cell lines on the HG-U133A GeneChip oligonucleotide microarray (Affymetrix, Santa Clara, CA) were obtained from previous publications [19,21–24]. The image file was analyzed with Robust Multichip Average to obtain the expression intensity values of the microarray data [25]. Reverse-phase protein microarrays (RPMAs), constructed from lysates of these 39 cell lines, were protein quantitated, stored, and blocked for immunodetection as described previously [26,27]. These arrays were then stained with antibodies recognizing total EGFR, and given its previous association with sensitivity to trastuzumab, HER2 pTyr-1248 [28], as well as 21 phosphospecific antibodies against signaling proteins in the EGFR pathway, using an automated slide stainer (Dako Cytomation, Carpinteria, CA) using the Catalyzed Signal Amplification System kit according to the manufacturer's recommendation (CSA; Dako Cytomation). Each antibody was extensively validated for single-band specificity and ligand induction by Western blot. The targets are listed in Figure 2D, whereas the sources, specificity workup, staining and detection procedures, and data analysis protocol for these antibodies have been reported recently [29]. This protein microarray platform has been demonstrated to be sensitive (detection levels approaching attogram quantities of target and variances of less than 10%) [30] and useful for analysis of patient tumor samples [31].
Figure 2.
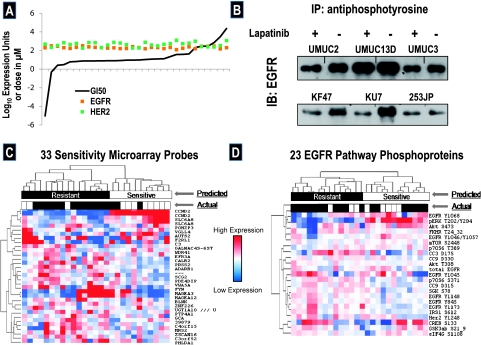
Lapatinib sensitivity compared with candidate biomarkers. (A) Cell lines were sorted by increasing lapatinib GI50 (concentration necessary to reduce cellular growth by one half relative to no drug) and their log10GI50s were plotted with their logged expression of the EGFR and HER2 (Affymetrix probe sets 211551_at and 216836_s_at). (B) The indicated cell lines, including those classified as sensitive (UMUC3), intermediate (KK47), and resistant (KU7, UMUC2 and 13D, and 253JP), were treated at their lapatinib GI50 or control for 24 hours before immunoprecipitation with anti-phosphotyrosine antibodies and detection with anti-EGFR antibodies. (C) Hierarchical clustering, as reported recently [63], of 27 (11 sensitive, 16 resistant) bladder cancer cell lines by 33 microarray probes significantly differentially expressed between them. The upper white and black panel shows a “predicted” split of the cell lines into two major clusters based on their expression of these genes, whereas the lower white (sensitive) and black (resistant) boxes show their actual in vitro determined sensitivity status. (D) A similar hierarchical cluster to panel (B) but clustering based on expression of 23 phosphoproteins in the EGFR pathway.
Bioinformatic and Statistical Analyses
For the cell lines used in the study, GI50s and GI30s were estimated based on in vitro dose-response curves fitted to a nonlinear regression model, as reported before [17,20]. As we [17,20] and others [32] have reported before, classification of cell lines as sensitive or resistant was done by the application of a criterion dose (CR) concept designed to classify based on both the 1) relevant biologic difference (sensitivity) and 2) ability to provide enough cell lines, approximately 10, in each class for robust statistical analysis of gene expression. Thus, we chose 8 µM as CR, defining cells as sensitive if GI50 < CR and resistant if GI30 > CR. Correlations of cell line's GI50s to microarray expression values of EGFR and HER2 were nonparametric Spearman correlations performed in GraphPad Prism Software (La Jolla, CA) and 2-tailed t tests and χ2 tests in Excel. For the development of prediction models, four discovery strategies were used. The first used HG-U133A (22,215 probes) microarray probes differentially expressed (significance analysis of microarrays [33] and local pooled error tests [34]) between sensitive and resistant cell lines. The second used the 23 EGFR-related phosphoproteins assayed by the RPMA platform. The third used a subset of the gene expression data, 60 EGFR pathway-relevant genomic expression biomarkers selected from the microarray based on the Kyoto Encyclopedia of Genes and Genomes database (www.genome.jp). The fourth strategy pooled both microarray gene expression and EGFR phosphoprotein activation data to generate and evaluate the relative contributions of the gene expression and EGFR pathway activation components to the efficacy of prediction.
Accurate, parsimonious prediction models for lapatinib sensitivity were then identified for the above four discovery strategies by the misclassification-penalized posterior (MiPP) method [35], which evaluates candidate prediction models based on both their accuracy and their strength of prediction. In brief, the MiPP algorithm first generates multiple independent splits of training and test data sets, which, in turn, result in multiple prediction models. Then, to evaluate the performance of the prediction models generated from this first step, the data set was split into test and training sets more than 100 times to generate a distribution of prediction scores for each model, from which their objective confidence bounds can be obtained. The performance of such models is described through use of the standardized MiPP score (sMiPP), which varies between -1 and 1, from the worst (low strength of prediction and least accuracy) to the best (high strength prediction and high accuracy). From this confidence interval evaluation, mean and lower 5% sMiPP scores (i.e., 95% likelihood that the model outperforms that value) and mean misclassification rates were obtained for each of the candidate prediction models. The MiPP algorithm is summarized in Figure W1.
Results
Sensitivity of Human Bladder Cell Lines to Lapatinib
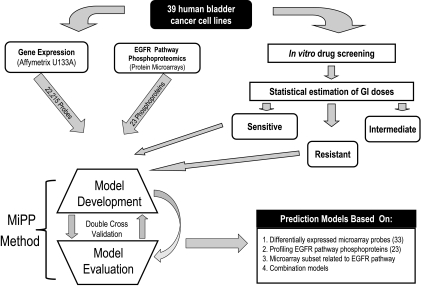
Our experimental algorithm for the development of molecular models predicting the sensitivity of bladder cancer cell lines to lapatinib is shown in Figure 1. The first step was to test in vitro the growth inhibitory activity of lapatinib in 39 bladder cancer cell lines. We generated classic dose-response curves, estimated GI50s using nonlinear regression analysis, and reported these in Table 1. To define cell lines as “sensitive” or “resistant” for downstream data analysis, we devised a criterion dose [17,20] as described in Materials and Methods. Eleven cell lines met our criterion for sensitivity, whereas 16 cell lines were determined to be resistant to lapatinib (Table 1). Cell lines that did not meet this criterion (i.e., had intermediate sensitivities) were excluded from further analysis (Figure W2).
Figure 1.
Experimental overview. Schematic overview for the development of prediction models for lapatinib sensitivity in human bladder cancer cell lines. Thirty-nine bladder cancer cell lines were subjected to in vitro determination of lapatinib sensitivity, baseline microarray gene expression profiling, and EGFR pathway phosphoprotein profiling by reverse-phase protein microarrays in parallel. Using these data for cell lines showing in vitro sensitivity and resistance to lapatinib through the MiPP algorithm, models were developed and evaluated based on all microarray data, EGFR pathway phosphoprotein data and a subset of array data for known EGFR pathway proteins.
Table 1.
Bladder Cell Lines, Lapatinib Sensitivity, and EGFR and HER2 Expression.
| Cell Line | Aliases | Sensitivity* | GI50 | GI30 | EGFR† | Her2‡ |
| 253J LAVAL | Sensitive | 9.46e - 06 | 1.51e - 17 | 197.662 | 475.005 | |
| UMUC6 | Sensitive | 0.472486 | 0.019145 | 312.573 | 222.158 | |
| 1A6 | Sensitive | 2.617867 | 0.376715 | 290.071 | 344.306 | |
| MGHU4 | Sensitive | 3.001725 | 0.779046 | 198.757 | 675.296 | |
| VMCUB2 | Sensitive | 6.047518 | 3.111507 | 165.954 | 430.082 | |
| 5637 | HTB-9 | Sensitive | 6.542723 | 3.393761 | 225.347 | 572.962 |
| Hs172.T | CRL-7833 | Sensitive | 7.537695 | 5.072047 | 320.731 | 180.246 |
| MGHU3 | Sensitive | 7.562033 | 5.280242 | 248.547 | 713.548 | |
| UM-UC-3 | CRL-1749 | Sensitive | 7.776003 | 6.140231 | 201.152 | 207.442 |
| SL4 | Sensitive | 7.898453 | 5.909908 | 180.938 | 385.1395 | |
| SW1710 | Sensitive | 7.9103 | 4.426662 | 194.459 | 746.945 | |
| T24T | Intermediate | 8.281807 | 3.704375 | 160.312 | 635.74 | |
| T24 | HTB-4 | Intermediate | 9.131583 | 7.233934 | 164.519 | 417.595 |
| KK47 | Intermediate | 9.319498 | 2.25336 | 254.375 | 183.997 | |
| UMUC3E | Intermediate | 9.543492 | 7.066357 | 204.852 | 187.281 | |
| VMCUB3 | Intermediate | 10.67859 | 5.603397 | 153.964 | 763.236 | |
| TCCSUP | HTB-5 | Intermediate | 11.75994 | 6.516546 | 215.506 | 385.619 |
| SCABER | HTB-3 | Intermediate | 12.92753 | 1.309457 | 158.343 | 465.638 |
| SW780 | CRL-2169 | Intermediate | 21.97725 | 4.588134 | 213.838 | 546.201 |
| UMUC9 | Intermediate | 37.4769 | 3.124082 | 122.157 | 1238.58 | |
| HT-1376 | CRL-1472 | Intermediate | 38.20096 | 5.763254 | 175.057 | 531.779 |
| CUBIII | Intermediate | 43.1401 | 1.203082 | 246.761 | 609.188 | |
| BC16.1 | Intermediate | 204.1396 | 6.68218 | 157.73 | 988.223 | |
| HT-1197 | CRL-1473 | Resistant | 12.42569 | 9.231061 | 209.47 | 311.749 |
| 253JP | Resistant | 288.4687 | 12.22306 | 281.861 | 248.071 | |
| 253JBV | Resistant | 288.1026 | 13.28975 | 282.737 | 260.826 | |
| PSI | Resistant | 694.7739 | 30.68547 | 302.276 | 239.202 | |
| UMUC14 | Resistant | 3016.783 | 33.70369 | 149.282 | 331.882 | |
| 575A | Resistant | 23,083.92 | 231.1572 | 200.782 | 1146.17 | |
| Hs228.T | CRL-7193 | Resistant | § | 330.749 | 231.088 | |
| FL3 | Resistant | § | § | 157.9835 | 573.9055 | |
| J82 | HTB-1 | Resistant | § | § | 163.089 | 477.636 |
| JON | Resistant | § | § | 225.689 | 1384.9 | |
| KU7 | Resistant | § | § | 256.618 | 202.81 | |
| RT4 | HTB-2 | Resistant | § | § | 240.453 | 1273.03 |
| UMUC1 | Resistant | § | § | 235.984 | 193.294 | |
| UMUC13D | Resistant | § | § | 221.113 | 167.805 | |
| UMUC2 | Resistant | § | § | 231.446 | 295.99 | |
| VMCUB1 | Resistant | § | § | 202.734 | 701.373 |
Cell line sensitivity was determined by the criterion dose 8 µM; sensitive cell lines, GI50 < 8 µM; resistant cell lines, GI30 > 8 µM. Cell lines were ranked within groups by GI50 (sensitive, intermediate) or GI30 (resistant).
Epidermal growth factor receptor, Affymetrix Probe ID: 211551_at.
HER2, ERBB2, Affymetrix Probe ID: 216836_s_at.
Resistant lines where growth curves were invariant across concentrations tested, precluding estimation of GI50 by regression analysis.
Evaluation of Gene Expression Prediction Models for Sensitivity to Lapatinib
Using published HG-U133A data for the 39 cell lines, we examined whether there was any relationship between the cell line's GI50s and the expression of lapatinib targets EGFR and HER2. Neither did we observe significant correlation between the cell line's GI50s and expression of these targets (rs = -0.1854, P = .33; rs = 0.08, P = .66, respectively; Figure 2A) nor did we observe a statistically significantly differential expression of these targets in cells defined as sensitive or resistant (P = .90 and P = .72, respectively). Given these findings, we wished to demonstrate that the drug was indeed functionally inhibiting its target; thus, we performed immunoprecipitation experiments with anti-phosphotyrosine antibodies and blot analysis with anti-total EGFR, finding variable inhibition of EGFR phosphorylation at cell lines' calculated GI50s (Figure 2B).
Next, we decided to use the entire microarray data to discover probes differentially expressed between sensitive and resistant cell lines. This analysis resulted in discovery of 33 probes significantly associated with lapatinib sensitivity (false discovery rate < 10%). Unsupervised two dimensional hierarchical clustering of these the 11 sensitive and 16 resistant cell lines by these 33 probes can be seen in Figure 2C, where each row represents the relative level of expression for the indicated probe (higher and lower expressions are shown as red or blue, respectively). We noted that clustering divided the cell lines into two large groups, roughly corresponding to sensitive and resistant clusters. Although the exchange of three sensitive and three resistant lines among these clusters rendered this split nonsignificant (P = .60), we interpreted the heat map as suggestive that predictive models might be built based on these probe sets.
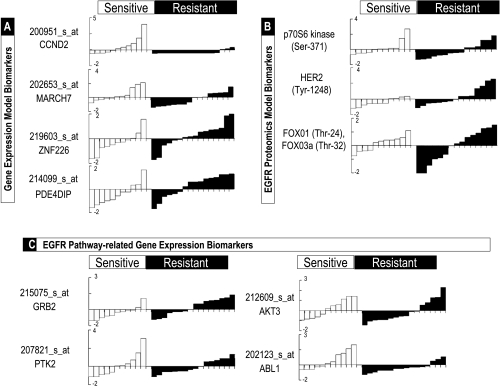
As reported before [17], we used the 33 sensitivity-related probes to construct prediction models of lapatinib sensitivity based on gene expression models using the MiPP technique [35]. Because of the function of the MiPP algorithm, which develops predictive models based on splitting training and test data (Figure W1), predictive models may contain some of the same or different genes (Table W1). A priori, we adopted the criterion that models evince at a minimum a lower 5% sMiPP score > 0.7 (i.e., 95% likelihood that the model performs better than an sMiPP of 0.7) and found that two predictive models met this criterion, Gene Expression Models 1 and 2, mean prediction accuracies 98.2% and 93.2%, respectively (Table 2). Gene Expression Model 1 included cyclin D2, membrane-associated ring finger (C3HC4) 7, and zinc finger protein 226. Gene Expression Model 2 was similar to Model 1, with the substitution of phosphodiesterase 4D interacting protein as the third prediction gene. The performance of these probes within these models across sensitive and resistant cell lines can be seen in Figure 3A.
Table 2.
Microarray Probe or Phosphoprotein-Specific Composition of the Five Predictive Models and Their Accuracy.
| Platform | Microarray Data | EGFR Pathway Genes | Reverse Phase Protein Microarray | Combination | |||
| Mean accuracy | 0.98 | 0.93 | 0.86 | 0.61 | 0.55 | 0.55 | 0.98 |
| Mean MiPP | 8.60 | 7.59 | 5.04 | 0.76 | 0.44 | 0.35 | 5.78 |
| Mean sMiPP | 0.96 | 0.84 | 0.63 | 0.19 | 0.11 | 0.09 | 0.96 |
| 5% sMiPP | 0.80 | 0.71 | 0.36 | -0.31 | -0.05 | 0.00 | 0.67 |
| 95% sMiPP | 1.00 | 0.98 | 0.84 | 0.67 | 0.33 | 0.35 | 0.99 |
| Biomarker 1 | CCND2 | CCND2 | GRB2 | p70S6 S371 | HER2 Y1248 | FOX01/03a T24/32 | FAM128B |
| Biomarker 2 | MARCH7 | MARCH7 | PTK2 | HER2 Y1248 | SLC6A8 | ||
| Biomarker 3 | ZNF226 | PDE4DIP | AKT3 | EGFR Y1173 | |||
| Biomarker 4 | ABL1 | ||||||
Figure 3.
MiPP models predictive of lapatinib sensitivity. (A) Two three-probe gene expression models were built based on a total of four probes for expression of the genes CCND2, MARCH7, ZNF226, and PDE4DIP. The sensitive (white) or resistant (black) cell lines' expression of each of these probes were z-scored (units of SD) for easy visualization and sorted before being plotted to show their differences of expression within sensitive and resistant groups. (B) Similar plots to panel (A) but of three EGFR pathway phosphoproteins contained in the three EGFR pathway models. (C) Similar plots to panels (A) and (B), but this time showing the expression patterns of four EGFR pathway-related gene expression probes comprising one model. Composition, accuracy, and MiPP performance parameters of these models are summarized in Table 2.
Evaluation of Proteomic Prediction Models
Because sensitivity to EGFR family inhibitors has also been correlated to the signaling status of this pathway [15], we sought to determine whether lapatinib sensitivity in our cell lines was associated with differential activation status. We used RPMA [26] to quantitatively interrogate the status of targets in this pathway. Thus, we spotted serial dilutions of the 39 cell line lysates onto nitrocellulose-coated glass slides and quantitated the activation/phosphorylation levels of 22 EGFR pathway phosphoproteins and total EGFR for each cell line, as described in Materials and Methods and reported before [29]. Unsupervised two dimensional hierarchical clustering of the 11 sensitive and 16 resistant cell lines by these 23 phosphoproteins can be seen in Figure 2D, clustered as in Figure 2C. Again, the clusters provided two main groupings of the cell lines, including one with many of the resistant cell lines, although compared with the microarray probes-based clusters, the EGFR-based groups showed greater divergence from actual in vitro sensitivity status.
The MiPP approach was used to develop sensitivity prediction models based on these EGFR pathway data. Whereas none of the models could satisfy our criterion of lower 5% sMiPP > 0.7, three top performing models were EGFR Proteomics Model 1, including the two phosphoprotein p70S6 kinase (pSer-371) and HER2 (pTyr-1248), mean accuracy 61%; EGFR Proteomics Model 2, including HER2 (pTyr-1248) alone, mean accuracy 55%; and EGFR Proteomics Model 3, including detection of FOX01 (pThr-24) and FOX03a (pThr-32) by a single antibody, mean accuracy 55% (Table 2 and Figure 3B).
Evaluation of Gene Expression Prediction Models from EGFR Pathway-Related Genes
Given our finding that proteomic prediction models based on EGFR pathway signaling status alone underperformed relative to those based on global gene expression, we were interested in evaluating models limited to gene expression data for components of the EGFR signaling pathway. We used the Kyoto encyclopedia of Genes and Genomes gene network database (www.genome.jp) to preselect 60 EGFR pathway-related genes for developing such prediction models. Using the same MiPP technique for discovery and evaluation of predictive models, we again found that there were no predictive models such that the lower 5% sMiPP score was higher than 0.7. The highest performance was shown by one model that included four probes and with a mean prediction accuracy of 86.2% (Table 2 and Figure 3C). Thus, transcriptional prediction based on models restricted to EGFR pathway also underperformed compared with models derived from unbiased genome-wide searches.
Evaluation of Combination Prediction Models
Because the predictive performance of nomograms can sometimes be enhanced by combination of several types of data [36], we were interested in whether a combination of both data sets (gene expression and phosphoproteomics) could provide equivalent or even improved predictive information beyond that provided by models derived from global transcriptional assessment. We used theMiPP algorithm to generate and evaluate combination models composed of both gene expression and EGFR pathway phosphoproteomics data. This was performed in an unbiased manner, pooling both data sets before application of the algorithm, which resulted in the discovery of three gene MiPP models that included one, two, or three probes from either data type. Then, top combination models were examined to evaluate the relative contributions of the gene expression probes versus EGFR phosphoproteins to predictive efficacy. We found that most of the top-performing models included only gene expression probes; however, the top-performing combination model included the phosphoprotein EGFR (pTyr-1173), with a mean prediction accuracy of 98.3%, similar to the gene expression-based models discovered based on differential expression between sensitive and resistant cell lines (Table 2).
Discussion
As more data become available from clinical trials of molecularly targeted drugs, it is increasingly clear that selection of patients that are likely to respond to such drugs needs to be undertaken a priori, to optimize the value of trials and discover patient subsets likely to benefit from treatment [37]. The development of response biomarkers, be they mutation, gene expression, proteomics-based, or, for that matter, a combination of these, is key in accomplishing this goal. Clinical trials done on patients with non-small cell lung cancer (NSCLC) and gefitinib highlight these points and demonstrate the utility of even a single biomarker in the prediction of patient response [38,39]. Recently identified somatic mutations in the tyrosine kinase domain of the EGFR gene in exons 18, 19, and 21 were identified in 18.9% of patients and found to be in a direct correlation with clinical response in these NSCLC patients to gefitinib. It was shown that 88.2% of patients with identified EGFR mutations experienced clinical benefit from treatment with gefitinib and that these mutations predicted not only response but also survival. On the basis of these results, the authors recommended testing patient tissues for such mutations and use as a first-line therapy in mutation-positive patients [36]. Recently, it was found that concomitant activation of Y1068 and Y1148 of the EGFR could predict mutation status of the gene in human lung cancer clinical specimens, revealing a common functional consequence of the known mutations on signaling activation [29].
However, no such EGFR mutations have been identified in bladder cancer cell lines and biopsy specimens [6], although a recent analysis of EGFR mutation in NCI-60 cell lines panel suggested that EGFR tyrosine kinase domain mutations may be rare in histologies other than NSCLC and potentially associated with resistance to inhibitors [40]. Also, although immunohistochemical markers based on known pathway targets have shown some promise in being found to be correlated with patient response to gefitinib [36] or even lapatinib [16], accurate prediction of response to these drugs, despite its importance to trial design and patient selection, remains a challenge.
To address the lack of lapatinib biomarkers in bladder cancer we used 1) genome-wide microarray data, 2) RPMA-based profiling of targets in the EGFR pathway, and 3) a subset of the microarray data relevant to the EGFR pathway to discover prediction models for lapatinib sensitivity across 39 bladder cancer cell lines. We found that a substantial proportion (11/39 or 28.2%) of bladder cancer cell lines exhibited sensitivity in vitro to lapatinib with GI50s lower than 8 µM. Although these values may initially appear higher than published studies [15], it is important to note that our studies were performed in RPMI with 10% serum (necessarily, to be able to grow all bladder cell lines in an internally consistent medium). Moreover, the proportion of cell lines classified as sensitive by application of the 8 µM criterion dose (∼25%) is similar to the proportion of patients showing clinical benefit from lapatinib in a very recently published clinical trial in advanced bladder cancer patients (∼35%) [41] and previous trials of lapatinib in breast cancer (∼18%) [42]. On the basis of this sensitivity classification scheme, we developed predictive algorithms based on comparing the aforementioned three types of molecular data between the 11 most sensitive and the 16 most resistant cell types.
We observed that predictive models based on 33 probes significantly differentially expressed between the 11 sensitive and the 16 resistant cell lines (mean accuracies across cross-validation loops, 93% and 98%) substantially outperformed models based on the EGFR pathway alone, whether it was interrogated through protein microarrays (mean accuracies, 55%, 55%, and 61%) or as a subset of the microarray data (mean accuracy, 86%). However, the potential to combine EGFR pathway data with gene expression data was demonstrated by our observation that EGFR pTyr-1173 could also contribute to a top-performing combination model (mean accuracy, 98%), supporting the importance of profiling phosphoprotein expression for biomarker discovery. Such combination strategies may, in the future, provide the opportunity for increased precision or robustness of prediction by combining data from different technologies and even traditional clinicopathologic parameters as has been noted in previous reports developing prognostic nomograms of bladder cancer [43,44].
Although we were intrigued that signatures developed based only on data from EGFR pathway targets underperformed, in retrospect, this may not be unexpected. One possible explanation is that, although the pathway mapping using RPMA produces a multiplexed view of cell signaling, the pathway signature in this study did not include every single known protein signaling end point of EGFR signaling, including several known direct downstream signaling targets of EGFR signaling such as SRC and SHC, STAT3, STAT5, as well as phosphospecific EGFR site Y992, and other EGFR family members, c-erbB3 and 4. Indeed, because broad-scale profiling of activation states of hundreds of signaling proteins is possible using RPMA, it may be possible to identify a pathway biomarker signature composed of other components of EGFR signaling that could provide a much enhanced predictive signature to lapatinib sensitivity. Such exploratory studies are underway.
Although two of the three phosphoproteomic models included HER2 pY1248, a site associated with activation [45], poor prognosis [46], therapeutic response to another anti-HER2 therapy (trastuzumab) breast cancer patients [28], and to be regulated in vitro [47] and in vivo [48] by lapatinib treatment, in this cohort of bladder cancer cell lines, the expression of the EGFR and HER2 protein targets themselves was not correlated to sensitivity to lapatinib (Figure 2). Consistent with that finding, in our previous reports, developing predictive signatures for combination sensitivity to the cytotoxic drugs, cisplatin, gemcitabine, and paclitaxel [17], we observed that most predictive probes did not have clear relationships to the drugs' known mechanism of action, suggesting the existence of novel pathways of drug response modulation.
Our gene expression models contained genes such as CCND2, MARCH7, ZNF226, and PDE4DIP, none of which, to our knowledge, have been associated with resistance to perturbation of EGFR family signaling before. Appearing in both of the models, cyclin D2 (CCND2) is a regulatory subunit of CDK4 and CDK6 kinases that are required for proper functioning of cell cycle G1/S transition, although related cyclin D1 expression has been shown to be modulated by lapatinib treatment in breast cancer cells [49]. Another gene appearing in both models was MARCH7 (membrane-associated ring finger (C3HC4)-7 or axotrophin). Although, as yet, uninvestigated in cancer, this gene is known to be involved in the regulation of the role of T lymphocytes in immune tolerance [50]. In the end, although our gene expression models do not contain biomarkers known to be associated with sensitivity to lapatinib, bladder cancer, which does not seem to harbor EGFR mutations [51] yet includes patient subsets who benefit from lapatinib therapy [41], may require such markers for optimal patient stratification.
Our findings come at a time of renaissance for cell lines as a bona fide platform for preclinical evaluation because of the development of predictive algorithms for patient tumors [52,53]. Two recent reports have shown the ability to predict, based on informatic approaches to gene expression and drug activity data in cell lines, the outcome of human clinical trials with available matched microarray data [20,54]. One of these algorithms, COXEN, also has the ability to perform in silico drug discovery, allowing generalization of novel agents in the development of specific tumor histologies of other tumor types based on core molecular similarities that transcend tissue-specific differences [20,52]. Thus, the data produced herein might, through such an approach, suggest investigation of lapatinib application for tumor types heretofore untried. Most compelling would be the application of our MiPP-based signatures and COXEN technology to attempt to predict clinical outcomes for a microarray data-matched lapatinib human clinical trial based on in vitro studies in our cell lines and other published cell line series [55,56]. At present, we are seeking such data for downstream validation of these signatures.
Whether such extrapolation is possible in the future, lapatinib represents an exciting new agent for bladder cancer. Not only do pathologic tissue studies [8,57,58] and in vitro models [10,11,59] suggest a significant role for EGFR and HER2 signaling in bladder cancer but also early clinical trial data in bladder cancer patients [60] suggest it might have activity in this histologic diagnosis. Indeed, a trial of lapatinib alongside the standard doublet, gemcitabine and cisplatin, as first-line therapy for locally advanced and metastatic EGFR or HER2 overexpressing bladder cancer, is currently in the recruiting stage (clinicaltrials.gov identifier: NCT00623064). Intriguingly, lapatinib-mediated reversal of drug resistance has been observed in other cell line models [61], whereas data from a new report suggest synergy between lapatinib and gemcitabine, paclitaxel, and cisplatin (a chemotherapeutic combination used in advanced disease) in a bladder cancer cell line [62]. Together, these exciting findings suggest that this agent deserves strong attention from investigators and trialists in urologic oncology.
Supplementary Material
Acknowledgments
The authors thank Neil Spector of the Duke University Department of Medicine for his guidance with experimental design. The authors also thank Alexander Baras of the UVA Pathology Department and all members of the Theodorescu laboratory for their helpful discussions.
Footnotes
This work was supported by the National Institutes of Health grant CA075115.
This article refers to supplementary materials, which are designated by Table W1 and Figures W1 and W2 and are available online at www.neoplasia.com.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Botteman MF, Pashos CL, Redaelli A, Laskin B, Hauser R. The health economics of bladder cancer: a comprehensive review of the published literature. Pharmacoeconomics. 2003;21:1315–1330. doi: 10.1007/BF03262330. [DOI] [PubMed] [Google Scholar]
- 3.Pashos CL, Botteman MF, Laskin BL, Redaelli A. Bladder cancer: epidemiology, diagnosis, and management. Cancer Pract. 2002;10:311–322. doi: 10.1046/j.1523-5394.2002.106011.x. [DOI] [PubMed] [Google Scholar]
- 4.Bellmunt J, Guillem V, Paz-Ares L, Gonzalez-Larriba JL, Carles J, Albanell J, Tabernero JM, Cortes-Funes H, Baselga J. Gemcitabine/paclitaxel-based three-drug regimens in advanced urothelial cancer. Eur J Cancer. 2000;36(Suppl 2):17–25. doi: 10.1016/s0959-8049(00)00081-2. [DOI] [PubMed] [Google Scholar]
- 5.von der Maase H, Sengelov L, Roberts JT, Ricci S, Dogliotti L, Oliver T, Moore MJ, Zimmermann A, Arning M. Long-term survival results of a randomized trial comparing gemcitabine plus cisplatin, with methotrexate, vinblastine, doxorubicin, plus cisplatin in patients with bladder cancer. J Clin Oncol. 2005;23:4602–4608. doi: 10.1200/JCO.2005.07.757. [DOI] [PubMed] [Google Scholar]
- 6.Villares GJ, Zigler M, Blehm K, Bogdan C, McConkey D, Colin D, Bar-Eli M. Targeting EGFR in bladder cancer. World J Urol. 2007;25:573–579. doi: 10.1007/s00345-007-0202-7. [DOI] [PubMed] [Google Scholar]
- 7.Sridhar SS, Seymour L, Shepherd FA. Inhibitors of epidermal-growth-factor receptors: a review of clinical research with a focus on non-small-cell lung cancer. Lancet Oncol. 2003;4:397–406. doi: 10.1016/s1470-2045(03)01137-9. [DOI] [PubMed] [Google Scholar]
- 8.Kiyoshima K, Oda Y, Kinukawa N, Naito S, Tsuneyoshi M. Overexpression of laminin-5 gamma2 chain and its prognostic significance in urothelial carcinoma of urinary bladder: association with expression of cyclooxygenase 2, epidermal growth factor receptor [corrected] and human epidermal growth factor receptor [corrected] 2. Hum Pathol. 2005;36:522–530. doi: 10.1016/j.humpath.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 9.Chow NH, Chan SH, Tzai TS, Ho CL, Liu HS. Expression profiles of ErbB family receptors and prognosis in primary transitional cell carcinoma of the urinary bladder. Clin Cancer Res. 2001;7:1957–1962. [PubMed] [Google Scholar]
- 10.Theodorescu D, Laderoute KR, Gulding KM. Epidermal growth factor receptor-regulated human bladder cancer motility is in part a phosphatidylinositol 3-kinase-mediated process. Cell Growth Differ. 1998;9:919–928. [PubMed] [Google Scholar]
- 11.Gildea JJ, Harding MA, Seraj MJ, Gulding KM, Theodorescu D. The role of Ral A in epidermal growth factor receptor-regulated cell motility. Cancer Res. 2002;62:982–985. [PubMed] [Google Scholar]
- 12.Medina PJ, Goodin S. Lapatinib: a dual inhibitor of human epidermal growth factor receptor tyrosine kinases. Clin Ther. 2008;30:1426–1447. doi: 10.1016/j.clinthera.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 13.Di Cosimo S, Baselga J. Targeted therapies in breast cancer: where are we now? Eur J Cancer. 2008;44:2781–2790. doi: 10.1016/j.ejca.2008.09.026. [DOI] [PubMed] [Google Scholar]
- 14.Meric-Bernstam F, Hung MC. Advances in targeting human epidermal growth factor receptor-2 signaling for cancer therapy. Clin Cancer Res. 2006;12:6326–6330. doi: 10.1158/1078-0432.CCR-06-1732. [DOI] [PubMed] [Google Scholar]
- 15.Rusnak DW, Alligood KJ, Mullin RJ, Spehar GM, Arenas-Elliott C, Martin AM, Degenhardt Y, Rudolph SK, Haws TF, Jr, Hudson-Curtis BL, et al. Assessment of epidermal growth factor receptor (EGFR, ErbB1) andHER2 (ErbB2) protein expression levels and response to lapatinib (Tykerb, GW572016) in an expanded panel of human normal and tumour cell lines. Cell Prolif. 2007;40:580–594. doi: 10.1111/j.1365-2184.2007.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnston S, Trudeau M, Kaufman B, Boussen H, Blackwell K, LoRusso P, Lombardi DP, Ben Ahmed S, Citrin DL, DeSilvio ML, et al. Phase II study of predictive biomarker profiles for response targeting human epidermal growth factor receptor 2 (HER-2) in advanced inflammatory breast cancer with lapatinib monotherapy. J Clin Oncol. 2008;26:1066–1072. doi: 10.1200/JCO.2007.13.9949. [DOI] [PubMed] [Google Scholar]
- 17.Havaleshko DM, Cho H, Conaway M, Owens CR, Hampton G, Lee JK, Theodorescu D. Prediction of drug combination chemosensitivity in human bladder cancer. Mol Cancer Ther. 2007;6:578–586. doi: 10.1158/1535-7163.MCT-06-0497. [DOI] [PubMed] [Google Scholar]
- 18.Balko JM, Potti A, Saunders C, Stromberg A, Haura EB, Black EP. Gene expression patterns that predict sensitivity to epidermal growth factor receptor tyrosine kinase inhibitors in lung cancer cell lines and human lung tumors. BMC Genomics. 2006;7:289. doi: 10.1186/1471-2164-7-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harding MA, Arden KC, Gildea JW, Gildea JJ, Perlman EJ, Viars C, Theodorescu D. Functional genomic comparison of lineage-related human bladder cancer cell lines with differing tumorigenic and metastatic potentials by spectral karyotyping, comparative genomic hybridization, and a novel method of positional expression profiling. Cancer Res. 2002;62:6981–6989. [PubMed] [Google Scholar]
- 20.Lee JK, Havaleshko DM, Cho H, Weinstein JN, Kaldjian EP, Karpovich J, Grimshaw A, Theodorescu D. A strategy for predicting the chemosensitivity of human cancers and its application to drug discovery. Proc Natl Acad Sci USA. 2007;104:13086–13091. doi: 10.1073/pnas.0610292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Titus B, Frierson HF, Jr, Conaway M, Ching K, Guise T, Chirgwin J, Hampton G, Theodorescu D. Endothelin axis is a target of the lung metastasis suppressor gene RhoGDI2. Cancer Res. 2005;65:7320–7327. doi: 10.1158/0008-5472.CAN-05-1403. [DOI] [PubMed] [Google Scholar]
- 22.Smith SC, Oxford G, Wu Z, Nitz MD, Conaway M, Frierson HF, Hampton G, Theodorescu D. The metastasis-associated gene CD24 is regulated by Ral GTPase and is a mediator of cell proliferation and survival in human cancer. Cancer Res. 2006;66:1917–1922. doi: 10.1158/0008-5472.CAN-05-3855. [DOI] [PubMed] [Google Scholar]
- 23.Gildea JJ, Seraj MJ, Oxford G, Harding MA, Hampton GM, Moskaluk CA, Frierson HF, Conaway MR, Theodorescu D. RhoGDI2 is an invasion and metastasis suppressor gene in human cancer. Cancer Res. 2002;62:6418–6423. [PubMed] [Google Scholar]
- 24.Oxford G, Smith SC, Hampton G, Theodorescu D. Expression profiling of Ral-depleted bladder cancer cells identifies RREB-1 as a novel transcriptional Ral effector. Oncogene. 2007;26:7143–7152. doi: 10.1038/sj.onc.1210521. [DOI] [PubMed] [Google Scholar]
- 25.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishizuka S, Charboneau L, Young L, Major S, Reinhold WC, Waltham M, Kouros-Mehr H, Bussey KJ, Lee JK, Espina V, et al. Proteomic profiling of the NCI-60 cancer cell lines using new high-density reverse-phase lysate microarrays. Proc Natl Acad Sci USA. 2003;100:14229–14234. doi: 10.1073/pnas.2331323100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheehan KM, Gulmann C, Eichler GS, Weinstein JN, Barrett HL, Kay EW, Conroy RM, Liotta LA, Petricoin EF., III Signal pathway profiling of epithelial and stromal compartments of colonic carcinoma reveals epithelialmesenchymal transition. Oncogene. 2008;27:323–331. doi: 10.1038/sj.onc.1210647. [DOI] [PubMed] [Google Scholar]
- 28.Hudelist G, Kostler WJ, Czerwenka K, Kubista E, Attems J, Muller R, Gschwantler-Kaulich D, Manavi M, Huber I, Hoschutzky H, et al. Her-2/neu and EGFR tyrosine kinase activation predict the efficacy of trastuzumab-based therapy in patients with metastatic breast cancer. Int J Cancer. 2006;118:1126–1134. doi: 10.1002/ijc.21492. [DOI] [PubMed] [Google Scholar]
- 29.VanMeter AJ, Rodriguez AS, Bowman ED, Jen J, Harris CC, Deng J, Calvert VS, Silvestri A, Fredolini C, Chandhoke V, et al. Laser capture microdissection and protein microarray analysis of human non-small cell lung cancer: differential epidermal growth factor receptor (EGPR) phosphorylation events associated with mutated EGFR compared with wild type. Mol Cell Proteomics. 2008;7:1902–1924. doi: 10.1074/mcp.M800204-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheehan KM, Calvert VS, Kay EW, Lu Y, Fishman D, Espina V, Aquino J, Speer R, Araujo R, Mills GB, et al. Use of reverse phase protein microarrays and reference standard development for molecular network analysis of metastatic ovarian carcinoma. Mol Cell Proteomics. 2005;4:346–355. doi: 10.1074/mcp.T500003-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Wulfkuhle JD, Edmiston KH, Liotta LA, Petricoin EF., III Technology insight: pharmacoproteomics for cancer—promises of patient-tailored medicine using protein microarrays. Nat Clin Pract Oncol. 2006;3:256–268. doi: 10.1038/ncponc0485. [DOI] [PubMed] [Google Scholar]
- 32.Coldren CD, Helfrich BA, Witta SE, Sugita M, Lapadat R, Zeng C, Barón A, Franklin WA, Hirsch FR, Geraci MW, et al. Baseline gene expression predicts sensitivity to gefitinib in non-small cell lung cancer cell lines. Mol Cancer Res. 2006;4:521–528. doi: 10.1158/1541-7786.MCR-06-0095. [DOI] [PubMed] [Google Scholar]
- 33.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jain N, Thatte J, Braciale T, Ley K, O'Connell M, Lee JK. Local-pooled-error test for identifying differentially expressed genes with a small number of replicated microarrays. Bioinformatics. 2003;19:1945–1951. doi: 10.1093/bioinformatics/btg264. [DOI] [PubMed] [Google Scholar]
- 35.Soukup M, Cho H, Lee JK. obust classification modeling on microarray data using misclassification penalized posterior. Bioinformatics. 2005;21(Suppl 1):i423–i430. doi: 10.1093/bioinformatics/bti1020. [DOI] [PubMed] [Google Scholar]
- 36.Han S-W, Kim T-Y, Hwang PG, Jeong S, Kim J, Choi IS, Oh D-Y, Kim JH, Kim D-W, Chung DH, et al. Predictive and prognostic impact of epidermal growth factor receptor mutation in non-small-cell lung cancer patients treated with gefitinib. J Clin Oncol. 2005;23:2493–2501. doi: 10.1200/JCO.2005.01.388. [DOI] [PubMed] [Google Scholar]
- 37.Simon R. The use of genomics in clinical trial design. Clin Cancer Res. 2008;14:5984–5993. doi: 10.1158/1078-0432.CCR-07-4531. [DOI] [PubMed] [Google Scholar]
- 38.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 39.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 40.Liu W, Wu X, Zhang W, Montenegro RC, Fackenthal DL, Spitz JA, Huff LM, Innocenti F, Das S, Cook EH, Jr, et al. Relationship of EGFR mutations, expression, amplification, and polymorphisms to epidermal growth factor receptor inhibitors in the NCI60 cell lines. Clin Cancer Res. 2007;13:6788–6795. doi: 10.1158/1078-0432.CCR-07-0547. [DOI] [PubMed] [Google Scholar]
- 41.Wulfing C, Machiels JP, Richel DJ, Grimm MO, Treiber U, DeGroot MR, Beuzeboc P, Parikh R, Petavy F, El-Hariry IA. A single-arm, multicenter, open-label phase 2 study of lapatinib as the second-line treatment of patients with locally advanced or metastatic transitional cell carcinoma. Cancer. 2009;115:2881–2890. doi: 10.1002/cncr.24337. [DOI] [PubMed] [Google Scholar]
- 42.Geyer CE, Forster J, Lindquist D, Chan S, Romieu CG, Pienkowski T, Jagiello-Gruszfeld A, Crown J, Chan A, Kaufman B, et al. Lapatinib plus capecitabine for HER2-positive advanced breast cancer. N Engl J Med. 2006;355:2733–2743. doi: 10.1056/NEJMoa064320. [DOI] [PubMed] [Google Scholar]
- 43.Shariat SF, Karakiewicz PI, Ashfaq R, Lerner SP, Palapattu GS, Cote RJ, Sagalowsky AI, Lotan Y. Multiple biomarkers improve prediction of bladder cancer recurrence and mortality in patients undergoing cystectomy. Cancer. 2008;112:315–325. doi: 10.1002/cncr.23162. [DOI] [PubMed] [Google Scholar]
- 44.Karam JA, Lotan Y, Karakiewicz PI, Ashfaq R, Sagalowsky AI, Roehrborn CG, Shariat SF. Use of combined apoptosis biomarkers for prediction of bladder cancer recurrence and mortality after radical cystectomy. Lancet. 2007;Oncol 8:128–136. doi: 10.1016/S1470-2045(07)70002-5. [DOI] [PubMed] [Google Scholar]
- 45.DiGiovanna MP, Carter D, Flynn SD, Stern DF. Functional assay for HER-2/neu demonstrates active signalling in a minority of HER-2/neu-overexpressing invasive human breast tumours. Br J Cancer. 1996;74:802–806. doi: 10.1038/bjc.1996.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cicenas J, Urban P, Kung W, Vuaroqueaux V, Labuhn M, Wight E, Eppenberger U, Eppenberger-Castori S. Phosphorylation of tyrosine 1248-ERBB2 measured by chemiluminescence-linked immunoassay is an independent predictor of poor prognosis in primary breast cancer patients. Eur J Cancer. 2006;42:636–645. doi: 10.1016/j.ejca.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 47.Zhang D, Pal A, Bornmann WG, Yamasaki F, Esteva FJ, Hortobagyi GN, Bartholomeusz C, Ueno NT. Activity of lapatinib is independent of EGFR expression level in HER2-overexpressing breast cancer cells. Mol Cancer Ther. 2008;7:1846–1850. doi: 10.1158/1535-7163.MCT-08-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spector NL, Xia W, Burris H, III, Hurwitz H, Dees EC, Dowlati A, O'Neil B, Overmoyer B, Marcom PK, Blackwell KL, et al. Study of the biologic effects of lapatinib, a reversible inhibitor of ErbB1 and ErbB2 tyrosine kinases, on tumor growth and survival pathways in patients with advanced malignancies. J Clin Oncol. 2005;23:2502–2512. doi: 10.1200/JCO.2005.12.157. [DOI] [PubMed] [Google Scholar]
- 49.Strecker TE, Shen Q, Zhang Y, Hill JL, Li Y, Wang C, Kim HT, Gilmer TM, Sexton KR, Hilsenbeck SG, et al. Effect of lapatinib on the development of estrogen receptor-negative mammary tumors in mice. J Natl Cancer Inst. 2009;101:107–113. doi: 10.1093/jnci/djn436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Metcalfe SM. Axotrophin and leukaemia inhibitory factor (LIF) in transplantation tolerance. Philos Trans R Soc Lond B Biol Sci. 2005;360:1687–1694. doi: 10.1098/rstb.2005.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blehm KN, Spiess PE, Bondaruk JE, Dujka ME, Villares GJ, Zhao Y-J, Bogler O, Aldape KD, Grossman HB, Adam L, et al. Mutations within the kinase domain and truncations of the epidermal growth factor receptor are rare events in bladder cancer: implications for therapy. Clin Cancer Res. 2006;12:4671–4677. doi: 10.1158/1078-0432.CCR-06-0407. [DOI] [PubMed] [Google Scholar]
- 52.Crunkhorn S. Anticancer drugs: forecasting drug responses. Nat Rev Drug Discov. 2007;6:782–783. [Google Scholar]
- 53.Overdevest JB, Theodorescu D, Lee JK. Utilizing the molecular gateway: the path to personalized cancer management. Clin Chem. 2009;55:684–697. doi: 10.1373/clinchem.2008.118554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Potti A, Dressman HK, Bild A, Riedel RF, Chan G, Sayer R, Cragun J, Cottrill H, Kelley MJ, Petersen R, et al. Genomic signatures to guide the use of chemotherapeutics. Nat Med. 2006;12:1294–1300. doi: 10.1038/nm1491. [DOI] [PubMed] [Google Scholar]
- 55.Greshock J, Cheng J, Rusnak D, Martin AM, Wooster R, Gilmer T, Lee K, Weber BL, Zaks T. Genome-wide DNA copy number predictors of lapatinib sensitivity in tumor-derived cell lines. Mol Cancer Ther. 2008;7:935–943. doi: 10.1158/1535-7163.MCT-07-2072. [DOI] [PubMed] [Google Scholar]
- 56.Wood ER, Truesdale AT, McDonald OB, Yuan D, Hassell A, Dickerson SH, Ellis B, Pennisi C, Horne E, Lackey K, et al. A unique structure for epidermal growth factor receptor bound to GW572016 (lapatinib): relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Res. 2004;64:6652–6659. doi: 10.1158/0008-5472.CAN-04-1168. [DOI] [PubMed] [Google Scholar]
- 57.Rajkumar T, Stamp GW, Pandha HS, Waxman J, Gullick WJ. Expression of the type 1 tyrosine kinase growth factor receptors EGFreceptor, c-erbB2 and c-erbB3 in bladder cancer. J Pathol. 1996;179:381–385. doi: 10.1002/(SICI)1096-9896(199608)179:4<381::AID-PATH603>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 58.Rotterud R, Fossa SD, Nesland JM. Protein networking in bladder cancer: immunoreactivity for FGFR3, EGFR, ERBB2, KAI1, PTEN, and RAS in normal and malignant urothelium. Histol Histopathol. 2007;22:349–363. doi: 10.14670/HH-22.349. [DOI] [PubMed] [Google Scholar]
- 59.Theodorescu D, Cornil I, Sheehan C, Man MS, Kerbel RS. Ha-ras induction of the invasive phenotype results in up-regulation of epidermal growth factor receptors and altered responsiveness to epidermal growth factor in human papillary transitional cell carcinoma cells. Cancer Res. 1991;51:4486–4491. [PubMed] [Google Scholar]
- 60.Wulfing C, Machiels JP, Richel D, Grimm MO, Treiber U, deGroot M, Beuzeboc P, Farrell J, Stone NL, Leopold L, et al. A single arm, multicenter, open label, phase II study of lapatinib as 2L treatment of pts with locally advanced/metastatic transitional cell carcinoma (TCC) of the urothelial tract. J Clin Oncol; 2005 ASCO Annual Meeting Proceedings; 2005. p. 4594. [Google Scholar]
- 61.Dai C-L, Tiwari AK, Wu C-P, Su X-D, Wang S-R, Liu D-G, Ashby CR, Jr, Huang Y, Robey RW, Liang Y-J, et al. Lapatinib (Tykerb, GW572016) reverses multidrug resistance in cancer cells by inhibiting the activity of ATP-binding cassette subfamily B member 1 and G member 2. Cancer Res. 2008;68:7905–7914. doi: 10.1158/0008-5472.CAN-08-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McHugh LA, Sayan AE, Mejlvang J, Griffiths TR, Sun Y, Manson MM, Tulchinsky E, Mellon JK, Kriajevska M. Lapatinib, a dual inhibitor of ErbB-1/-2 receptors, enhances effects of combination chemotherapy in bladder cancer cells. Int J Oncol. 2009;34:1155–1163. doi: 10.3892/ijo_00000244. [DOI] [PubMed] [Google Scholar]
- 63.Saal LH, Johansson P, Holm K, Gruvberger-Saal SK, She Q-B, Maurer M, Koujak S, Ferrando AA, Malmström P, Memeo L, et al. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proc Natl Acad Sci USA. 2007;104:7564–7569. doi: 10.1073/pnas.0702507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.