Figure 1.
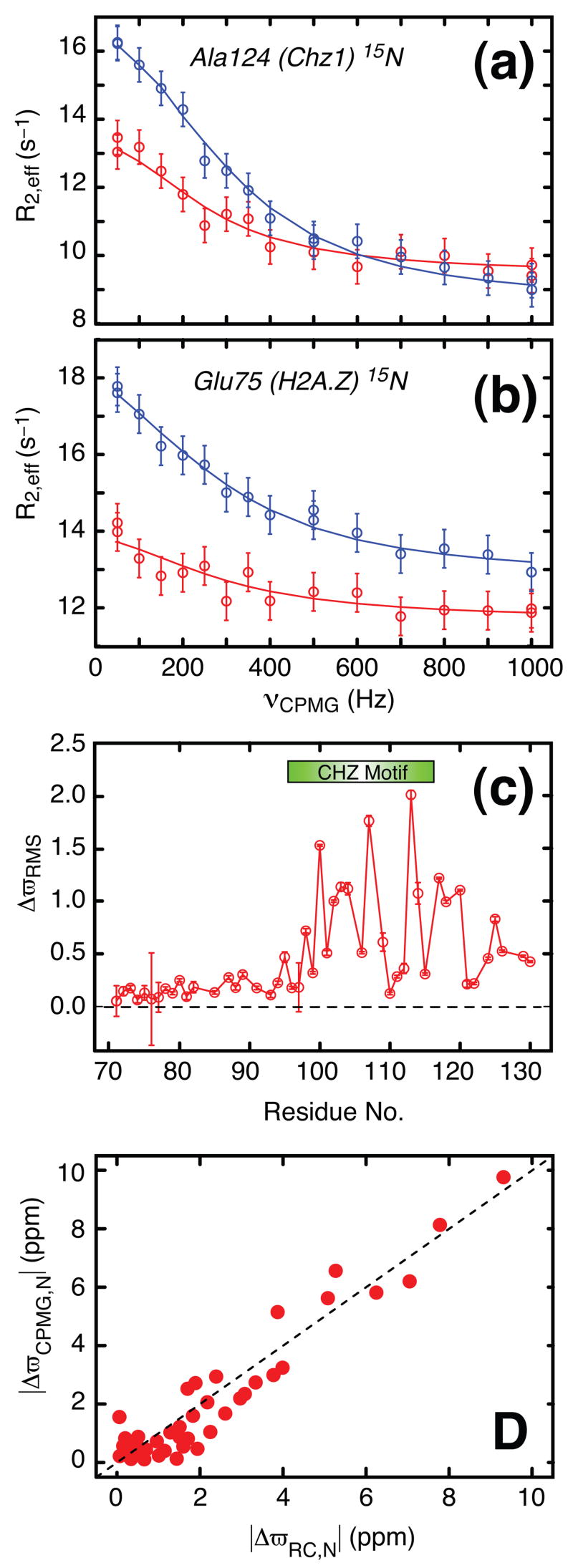
Amide 1H and 15N relaxation dispersion profiles have been recorded on a complex comprising the single stranded H2A.Z-H2B protein and the chaperone Chz.core (residues 71–132 of Chz1), prepared as described previously17. Both protein constituents were U-15N, 2H labeled. The NMR sample was prepared with [H2A.Z-H2B]Total≈1 mM, [Chz1]Total ≈1 mM and 25 mM MES, 0.2 M NaCl, 1 mM EDTA, 10% D2O, pH = 6.0. Representative 15N relaxation dispersion profiles from Ala124 of Chz1 (a) and Glu 75 of H2A.Z (b) recorded at static magnetic field strengths of 18.8 T (blue) and 11.7 T (red), 35°C are shown. The vertical lines associated with each measurement (circle) are error estimates; error values of 1% of R2, 0.3 s−1 or the standard deviation (SD) of duplicate measurements (whichever is the largest) were assigned to the 15N rates, while errors in 1HN transverse relaxation rates were based on maximum(3% of R2, 0.75 s−1, SD of duplicate measurements). The solid lines are fits of a two-site exchange model to the data: |Δϖ|=2.3±0.1 ppm for Ala124(Chz), |Δϖ|=1.8±0.1ppm for Glu75(H2A.Z). Exchange rates and populations are given in the text. (c) Residue specific ΔϖRMS values calculated as described in the legend to Figure 2. The CHZ motif (residue 95–115) is highlighted with a green bar. (d) Correlation plot of 15N chemical shift differences for Chz1 measured in a CPMG relaxation dispersion experiment, ΔϖCPMG,N, versus the difference between the assigned chemical shifts of the bound state of Chz1 and random roil values27, ΔϖRC,N. Relaxation dispersion profiles were recorded using Varian Inova NMR spectrometers operating at static magnetic field strengths of 11.7 T and 18.8 T (500 and 800 MHz 1H frequency), respectively. Constant-time22 relaxation compensated20 TROSY-based pulse schemes were used for recording 1H 34 and 15N 21 relaxation dispersion profiles. A constant-time relaxation delay of 25 ms (40 ms) was used for the proton (nitrogen) dispersion profiles. Effective 15N transverse relaxation rates (R2,eff) were measured for 14 different νCPMG frequencies between 50 and 1000 Hz, while the proton dispersion profiles were sampled at 15 different νCPMG frequencies between 80 and 1840 Hz. Data sets were processed with the NMRPipe program35 and signal intensities quantified using the program FuDA (flemming@pound.med.utoronto.ca; http://pound.med.utoronto.ca/software). Relaxation dispersion profiles, R2,eff(νCPMG), were generated from peak intensities, I(νCPMG), in a series of 2D 1HN-15N correlation maps measured as a function of CPMG frequency (νCPMG=1/2τ, where τ is the time between two successive refocusing pulses). Peak intensities were converted into effective relaxation rates via R2,eff= ln[I0/I(νCPMG)]/Trelax, where I0 is the peak intensity in a reference spectrum recorded without the constant-time relaxation delay Trelax. Details of the procedure by which dispersions profiles are fit are provided in the legend to Figure 3.
