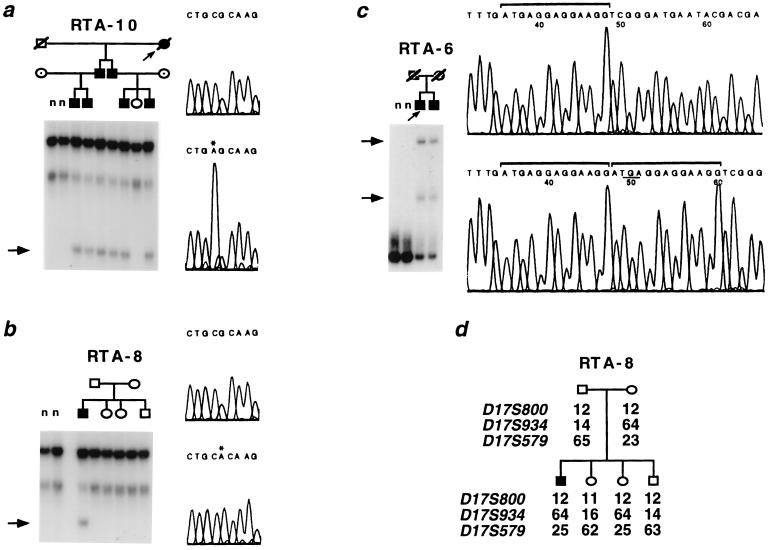
Figure 4.
Mutations in AE1 in dominant dRTA patients. The structure of each kindred is shown. Affected, unaffected, and phenotype-unknown subjects are indicated by solid, open, and shaded symbols, respectively. n = unrelated normal subjects. Below the diagram of each kindred in a–c are the results of single-strand conformational polymorphism analysis of exons 14, 14, and 20 of AE1, respectively. The corresponding DNA sequence of the sense strand of wild-type (Upper) and mutant alleles (Lower) are also shown. Arrows indicate variants specific to RTA patients. In a and b, asterisks above sequences indicate the variant bases in codon Arg-589. (a) In kindred RTA10, CGC is changed to AGC (589Ser) in all six affected members. (b) In kindred RTA8, only the affected index case shows the de novo mutation altering CGC to CAC (589His). (c) The bracket shows the 13-bp sequence present in single copy in wild-type sequence, but duplicated in tandem in both affected members of RTA6. The result leads to premature termination at codon 901 (underlined), truncating the protein by 11 amino acids. (d) Genotypes and haplotypes of kindred RTA8. Haplotypes flanking AE1 are shown, which confirmed biological parentage, and in addition show that the index case was identical with an unaffected sister. The results confirm the presence of a de novo mutation.