Figure 5.
Analysis of the MYB305 Activation Domain.
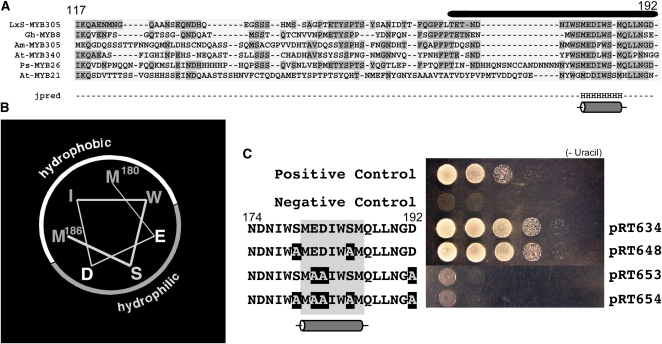
(A) Alignment of amino acid sequences of proteins closely related to LxS-MYB305. The amino acid numbers in LxS-MYB305 are indicated above the sequences, and the bar represents the minimal activation domain identified in Figure 4. Regions of identity are shaded. The secondary structural predictor Jpred predicts only one region of secondary structure, a single α-helix located in the center of the minimal activation domain.
(B) Surface analysis of the predicted α-helix showing the hydrophobic and hydrophilic surfaces. The positions of Met-180 and Met-186 are identified.
(C) Site-specific mutagenesis of the minimal activation domain of the LxS-MYB305. Serial dilution growth assays of transformed yeast on selective YSM Ura− medium. Effector and reporter constructs are identical to those in Figure 4. Positive (Gal4DB-AD) and negative (pDBleu) controls are shown. The site-specific mutants are identified to the left of the figure, with construct numbers shown at right. The location of the predicted α-helix is shaded and shown below these sequences. The spots are serial dilutions of each transformed yeast strain.
[See online article for color version of this figure.]