Figure 1.
Isolation of Arabidopsis rpoTmp Mutants.
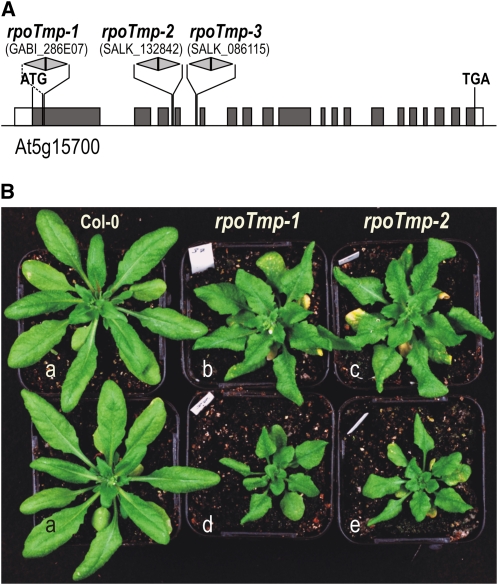
(A) RPOTmp gene organization and positions of T-DNA insertions in the three rpoTmp mutant alleles rpoTmp-1, rpoTmp-2, and rpoTmp-3. RPOTmp gene exons are represented as boxes (dark gray, coding sequences; white, untranslated regions). Gray arrowheads indicate T-DNA insertions and point toward the T-DNA left border. Note that double arrowheads represent two T-DNA copies inserted as inverted repeats, with left borders facing outward. Through PCR with primers annealing to the T-DNA left border and the flanking RPOTmp sequence, all insertions were found to be present as inverted repeats of the T-DNA. Insertion sites were verified by sequencing of PCR products.
(B) Phenotypes of wild-type plants (Col-0, plants labeled a) and rpoTmp-1 (b and d) and rpoTmp-2 mutant plants (c and e) grown in a 16-h photoperiod. A comparison of 4.5-week-old wild-type and mutant plants (a, d, and e) illustrates the developmental delay of rpoTmp mutants. Mutants grown for 6 weeks (b and c) are approximately at the same developmental stage and of a similar size as 4.5-week-old wild-type plants.