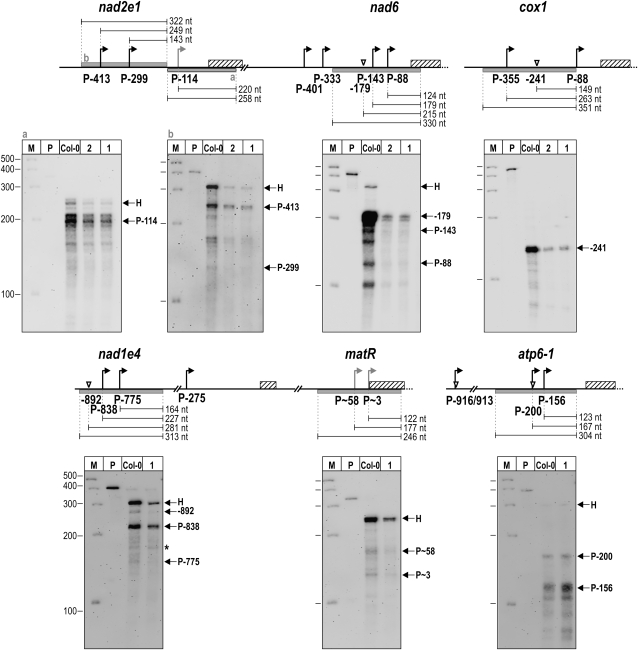
Figure 7.
Transcript 5′ End Abundances in rpoTmp and Wild-Type Seedlings.
RPAs were performed on total seedling RNA prepared from rpoTmp-1 (lanes labeled 1), rpoTmp-2 (2), and the wild type (Col-0) using biotinylated riboprobes annealing to the nad2e1, nad6, cox1, nad1e4, matR, and atp6-1 5′ regions. Protected RNA fragments were separated on polyacrylamide gels alongside a molecular weight marker (lane M) and the probe (P) used in the respective assay; sizes are given in nucleotides. Specific protected fragments are indicated by arrows and labeled with the name of the promoter or processing site they correspond to. Signals marked H are derived from protected fragments corresponding to the complete homologous segment of a probe. An asterisk in the nad1e4 panel marks a signal that was considered nonspecific, as it was seen with nonspecific RNA in a control assay (data not shown). Diagrams of the nad2e1, nad6, cox1, nad1e4, matR, and atp6-1 5′-untranslated regions above each gel image illustrate positions of promoters (bent arrows) and processing sites (triangles); coding sequences are shown as hatched bars. Gray bars are drawn below sequences complementary to the riboprobes that were used in RPAs. The sizes of expected protected RNA fragments are given in nucleotides. The two protected fragments derived from the nad2e1 probe “a” migrated slightly faster than expected, possibly due to secondary structure formation. A fragment that could correspond to PmatR∼3 appears slightly larger than expected.