Abstract
The aryl hydrocarbon receptor (AHR) is a widely expressed ligand-dependent transcription factor that mediates cellular responses to dioxins and other planar aromatic hydrocarbons. Ahr-null mice are refractory to the toxic effects of dioxin exposure. Although some mechanistic aspects of AHR activity are well understood, the tissue specificity of AHR effects remains unclear, both during development and following administration of exogenous ligands. To address the latter issue, we defined and compared transcriptional responses to dioxin exposure in the liver and kidney of wild-type and Ahr-null adult C57BL/6J mice treated with either 2,3,7,8-tetrachlorodibenzo-p-dioxin or corn-oil vehicle. In both tissues, essentially all effects of dioxin on hepatic mRNA levels were mediated by the AHR. Although 297 genes were altered by dioxin exposure in the liver, only 17 were changed in the kidney, including a number of well-established AHR target genes. Ahr genotype had a large effect in both tissues, profoundly remodeling both the renal and hepatic transcriptomes. Surprisingly, a large number of genes were affected by Ahr genotype in both tissues, suggesting the presence of a basal AHR gene battery. Alterations of the renal transcriptome in Ahr-null animals were associated with perturbation of specific functional pathways and enrichment of specific DNA motifs. Our results demonstrate the importance of intertissue comparisons, highlight the basal role of the AHR in liver and kidney, and support a role in development or normal physiology.
Keywords: aryl hydrocarbon receptor, dioxin, TCDD, microarray, kidney, liver
The aryl hydrocarbon receptor (AHR) is a ligand-dependent transcription factor, widely expressed in vertebrate tissues, that mediates cellular responses to dioxins and related planar aromatic hydrocarbons (Okey, 2007). The AHR resides quiescent in a cytoplasmic complex containing HSP90 and several other components (Hankinson, 1995). Upon ligand-binding the AHR translocates into the nucleus where it exists as a dimer with the AHR nuclear translocator (ARNT, also known as HIF1B) (Hankinson, 2005; Matsushita et al., 1993). The AHR:ARNT:ligand complex associates directly with DNA in a sequence-specific fashion, modulating the expression of target genes through the recruitment or interference of other regulatory proteins, such as transcription factors (Kobayashi et al. 1996; Ohtake et al. 2003; Sogawa et al. 2004), coactivators (Beischlag et al., 2002; Nguyen et al., 1999), and components of the basal transcriptional machinery (Rowlands et al., 1996).
The transcriptional effects of AHR activation are of profound importance in several biological systems. Mice whose Ahr locus has been genetically ablated are viable, but suffer several developmental defects (Schmidt et al., 1996; Walisser et al., 2004). Further, they are essentially immune to most toxicities induced by AHR ligands, and in particular the prototypical ligand 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) (Lin et al., 2008; Schmidt et al., 1996; Walisser et al., 2004). AHR activity has recently been shown to be essential for the development of regulatory T cells (Quintana et al., 2008; Veldhoen et al., 2008).
Several gaps prevent us from linking our mechanistic knowledge of AHR function to the many and varied phenotypic consequences of AHR activity. One of these is a limited understanding of the tissue specificity of AHR target genes, both during development and following stimulation by exogenous ligands. Although the transcriptional effects of AHR activity in liver are relatively well characterized (Boverhof et al., 2006; Fletcher et al., 2005; Franc et al., 2008; Frueh et al., 2001; Moennikes et al., 2004; Puga et al., 2000), other organs have received less attention. As a result, the degree of intertissue conservation of AHR target genes, binding motifs, and functional pathways is unknown.
Although the liver is well characterized as a site of major TCDD-induced toxicities, the kidney also plays an important role. During fetal development in mice, kidney is one of the most susceptible target tissues for TCDD, displaying hydronephrosis at doses that are too low to cause cleft palate (Couture et al., 1990). The kidney change is secondary to induced proliferation of ureteral epithelium by TCDD associated with an increase in EGF receptor density (Abbott and Birnbaum, 1990; Abbott et al., 1987). Overall, rats are more resistant to this impact of TCDD, but there are marked interstrain differences (Huuskonen et al., 1994).
We previously demonstrated that adult mice whose AHR locus has been genetically ablated (Ahr−/− mice) exhibit profoundly altered hepatic transcriptomes, even in the absence of TCDD exposure (Tijet et al., 2006). Here we describe, for the first time, the transcriptional effects of Ahr knockout on adult kidney, both in the presence and absence of TCDD treatment. Although the kidney transcriptome is mostly refractory to TCDD exposure, genetic ablation of AHR expression profoundly alters mRNA levels.
MATERIALS AND METHODS
2,3,7,8-Tetrachlorodibenzo-p-dioxin.
TCDD was purchased from the UFA-Oil Institute (Ufa, Russia) and was > 99% pure as determined by gas chromatography-mass spectrometry.
Animals and in vivo treatment.
Male, Ahr-null (Ahr−/−) mice 10 weeks of age were obtained from The Jackson Laboratory (Bar Harbor, ME). Male, wild-type (Ahr+/+) C57BL/6 mice 15 weeks of age were bred at the National Public Health Institute, Kuopio, Finland, from stock originally obtained from The Jackson Laboratory. Mice were handled as described previously (Tijet et al., 2006). Briefly, a single dose of 1000 μg/kg TCDD or corn-oil vehicle was given by gavage, and liver and kidney were harvested 19 h after treatment, sliced, snap frozen in liquid nitrogen and stored at −80°C until homogenization. All animal study plans were approved by the Animal Experiment Committee of the University of Kuopio and the Provincial Government of Eastern Finland. Twelve wild-type Ahr+/+ mice were used: six treated with TCDD and six treated with corn-oil vehicle. Six Ahr-null mice were used: three treated with TCDD and three treated with corn-oil vehicle.
RNA isolation.
Total RNA was extracted using RNeasy kits (Qiagen, CA) according to the manufacturer's instructions. DNase (Qiagen) was added to the RNeasy elution column as recommended by the manufacturer. RNA yield was quantified by UV spectrophotometry and RNA integrity was verified using an Agilent 2100 BioAnalyzer. The isolated RNA was then assayed on Affymetrix MOE430-2 arrays at The Centre for Applied Genomics at The Hospital for Sick Children (Toronto, Canada) following standard manufacturer's protocols. Each tissue was hybridized to a separate array, but one wild-type vehicle-treated liver sample was not run, leading to 35 total arrays.
Preprocessing of microarray data.
Array data were loaded into the R statistical environment (v2.8.1; R Development Core Team, 2008) using the affy package (v1.20.2) of the BioConductor open-source library (Gautier et al., 2004). Array data were investigated for spatial and distributional homogeneity; all arrays were included in subsequent analyses. The liver and kidney data were separately preprocessed using the GCRMA algorithm (Wu et al., 2004), as implemented in the gcrma package (v2.14.1) for the R statistical environment (v2.8.1) (R Development Core Team, Vienna, Austria). The array data were associated with updated sequence annotation using the mouse4302mmentrezg package (v11.0.1), which re-maps each 25-bp probe to a specific Entrez Gene ID (Dai et al., 2005). Raw and preprocessed microarray data are available in the GEO repository (accession GSE15859).
Statistical analysis of microarray data.
Within each tissue the experiment has a two-factor, two-level design with AHR status (knockout or wild-type) and TCDD treatment (vehicle or TCDD) as the factors. Accordingly we fit a general-linear model (GLM) using the limma package (v2.16.4) in BioConductor, as previously (Tijet et al., 2006). The following linear model was fit to each ProbeSet:
Here Y refers to the expression level of a single ProbeSet; Basal refers to the underlying basal expression level across all animals; AHR captures “AHR-dependent, TCDD-independent” expression changes; TCDD captures “TCDD-dependent, AHR-independent” expression changes; and AHR:TCDD captures “AHR-dependent, TCDD-dependent” expression changes. This linear model was fit separately to the preprocessed liver and the kidney data and the results subjected to an empirical Bayes moderation of the standard error (Smyth, 2003), followed by a false-discovery rate correction for multiple testing (Efron and Tibshirani, 2002). The significance threshold was set at padjusted < 0.05 for all comparisons.
Clustering of microarray data.
Unsupervised machine-learning employed the DIANA divisive hierarchical clustering algorithm, as implemented in the cluster package (v1.11.12) of the R statistical environment (v2.8.1). Pearson's correlation coefficient was used as the distance metric, and scaling was performed before clustering as previously (Boutros et al., 2008).
Functional enrichment analyses.
To analyze the functional pathways associated with Ahr genotype and the AHR-dependent response to TCDD in liver and kidney, we employed Gene Ontology (GO) enrichment analysis. Six gene lists were created: the liver-specific, kidney-specific, and common genes displaying responses to either Ahr genotype or to the AHR:TCDD interaction. For each gene on this list GO terms were identified, and for each GO term statistical significance was assessed using Fisher's Exact test, as implemented in the GOMiner tool (Zeeberg et al., 2003). All mouse GO databases available from GOMiner were used in this analysis. All three GO ontologies and all evidence codes were included. We used 1000 permutations to estimate the false-discovery rate. The resulting list of p values was log10 transformed and genes with pcumulative < 10−12 (i.e., a sum of the log10(p values) across the six lists smaller than 12 were extracted. This matrix was subjected to unsupervised hierarchical clustering using the DIANA algorithm with Pearson's correlation as the distance metric, as described above.
Transcription factor binding-site analyses.
We performed a targeted analysis of three transcription factor binding-site motifs known to be associated with transcriptional regulation by the AHR: the aryl hydrocarbon response elements I and II (AHRE-I and AHRE-II) and the antioxidant response element (ARE). We employed established methods to identify these motifs in the same 6001 bp of regulatory DNA as assessed in the library-based search (Boutros et al., 2004). Evolutionary conservation was assessed using the 30-vertebrate species PhastCons alignment scores from the UCSC Genome Browser Database (Siepel and Haussler, 2004), as described previously (Moffat et al., 2007). The results were visualized using violin plots with the lattice package (v0.17-22) for the R statistical environment (v2.8.1) and default parameters.
To identify transcription factors putatively enriched in the regulatory regions of genes whose mRNA abundances were altered by TCDD exposure (AHR:TCDD) or by Ahr genotype (AHR) we employed a library-based analysis using the CLOVER motif-finder algorithm (Frith et al., 2004) and the JASPAR open-source motif library (Sandelin et al., 2004). We searched 6001 bp of regulatory sequence, centered on the transcriptional start-site and extracted from the UCSC genome assembly (build mm9) using the RefSeq annotation in the UCSC genome browser (Karolchik et al., 2003) as downloaded on 2009-03-24. CLOVER was configured to return all motifs with p values less than 0.5 and motif-scores greater than three, and three separate statistical tests were performed. First, standard dinucleotide randomization was employed. Second, a background file containing the regulatory regions of all mouse genes was used. Third, a background file containing the regulatory regions of all mouse genes contained on our microarray platform was used. Only transcription-factor binding sites (TFBS) motifs that were enriched at p < 0.05 in all three statistical tests were considered. This analysis was repeated separately for the promoter regions of genes whose abundances were altered by TCDD exposure or Ahr genotype in the kidney alone, the liver alone, or both organs. The results were visualized using unsupervised machine-learning as described above.
Real-time RT-PCR validation.
Validation of the microarray data was performed by two-step real-time PCR analysis. cDNA derived from 100 ng of RNA was used for each real-time PCR reaction. Reverse transcription of RNA to cDNA was carried out with random hexamers and Moloney murine leukemia virus (MMLV) reverse transcriptase. Annealing of the random hexamers to the mRNA template was performed at 65°C for 5 min, followed by cooling to room temperature for 10 min. The extension reaction in the presence of MMLV was carried out at 37°C for 50 min, and was terminated by incubating the samples at 70°C for 10 min.
Relative quantitation of gene expression was performed by real-time PCR analysis on an ABI7700 sequence detection system (Applied Biosystems, Inc., Foster City, CA) using TaqMan gene expression assays (Applied Biosystems, Inc.). All TaqMan primer and probe sets contained Minor Groove-Binder (MGB) probes with the 5′ label dye FAM and a 3′ nonfluorescent quencher, with the exception of the assays for H+K+ATPase, Cd44, and Cyp1a1, which used Tetramethyl-6-Carboxyrhodamine (TAMRA) dye as a quencher. Beta-actin (Actb) served as the housekeeping gene for both sets of assays. The appropriate beta-actin assay (MGB-non fluorescent quencher or TAMRA quencher) was used in conjunction with the assay for a particular target gene.
Relative quantitation of target gene expression in relation to beta-actin (Actb) was determined from the mean normalized expression (Muller et al., 2002; Simon, 2003), which takes the mean threshold cycle (Ct) and the amplification efficiency of both the target and the housekeeping genes into account . The amplification efficiency of each Taqman gene expression assay was verified by constructing a standard curve that spanned a six-log dilution range of the target cDNA (log|concentration|) versus the corresponding Ct (threshold cycle) value for each sample; the dilution range encompassed the quantity of input cDNA (100 ng) for each target. The amplification efficiency of each assay was calculated from the slope of the line (Applied Biosystems Literature).
For each gene, three animals were assayed from each biological condition (12 animals in total) and the real-time PCR reaction was performed in duplicate for each animal. For TaqMan MGB assays, each reaction (one well in a 96-well plate) consisted of 10 μl of TaqMan Universal PCR Master Mix (2×), 7 μl of H2O, 1 μl of the Primer/Probe, and 2 μl (100 ng) of cDNA in a total reaction volume of 20 μl. For each custom assay, the reaction consisted of 10 μl of TaqMan Universal PCR Master Mix (2×), 2 μl of forward primer, 2 μl of reverse primer, 2 μl of the TaqMan probe, and 20 ng of cDNA. Primer and probe concentrations for the custom assays were optimized by running real-time PCR reactions with different primer or probe concentrations, whereas all other components remained constant. The primer and probe concentrations that yielded the lowest Ct (cycle threshold value) and the highest delta Rn (difference in the fluorescence emission intensity between a completed reaction and an unreacted sample) were chosen as optimal (Applied Biosystems Literature). Thermal cycling parameters encompassed an initial 20-s denaturation step at 95°C followed by 40 cycles of melting (1 s at 95°C) and annealing/extension (20 s at 60°C). Statistical analysis of PCR data was performed according to the identical linear-modeling procedure used for the microarray data, with the exception that an adjustment for multiple testing was not necessary (n = 11).
RESULTS
Experimental Design
We used mRNA transcript microarrays to study the transcriptional response to 1000 μg/kg TCDD or corn-oil vehicle in adult Ahr−/− and Ahr+/+ mice (Fig. 1). Animals were sacrificed nineteen hours after exposure, a time point around or prior to early manifestations of hepatic toxicity in mice (Boverhof et al., 2006). Kidneys and livers were excised and subjected to mRNA transcriptional profiling on oligonucleotide microarrays. The dose chosen corresponds to approximately three-fold the individual LD50 for male Ahr+/+ C57/BL6 animals (Pohjanvirta, 2009). Taken together, then, we expect this choice of dose and time point to allow identification of the early and acute results of dioxin exposure. The hepatic transcriptional response was reported previously (Tijet et al., 2006).
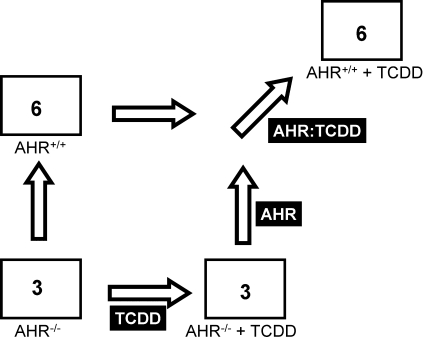
FIG. 1.
Experimental design. Mice harboring either the wild-type AHR (i.e., AHR+/+) or with genetic ablation of the AHR locus (i.e., AHR−/−) in a C57BL/6J background were treated with either corn-oil vehicle (not indicated) or 1000 μg/kg TCDD (i.e., +TCDD). The number of mice used was six in AHR+/+ conditions and three for the AHR−/− conditions. Both liver and kidney were excised 19 h after treatment, RNA extracted, and microarray profiling of mRNA abundances performed as described in “Material and Methods.” General-linear modeling was used to identify three effects. The AHR effect (labeled as “AHR”) gives alterations in mRNA abundance dependent on Ahr genotype, even in the absence of TCDD exposure. The TCDD effect (labeled as “TCDD”) gives alterations in mRNA abundance dependent on TCDD exposure, even in AHR−/− animals. Finally the AHR-TCDD interaction (labeled as “AHR:TCDD”) gives effects dependent on both Ahr genotype and TCDD exposure.
To ensure that transcriptional profiles from our new kidney data and older liver data are directly comparable we employed the same microarray platform and hybridization protocols for the two studies (Affymetrix MOE430-2 arrays) and analyzed the datasets using identical algorithms and software versions. All microarray data were carefully assessed for quality; none were excluded. Raw and preprocessed microarray data have been deposited in the GEO archive (accession GSE15859).
Transcriptional Profiling of Kidney
Using a GLM, we identified genes whose mRNA abundances were altered by Ahr genotype alone, by TCDD exposure alone, or by TCDD exposure in an AHR-dependent manner. To make accurate comparisons with our previous liver data, this same analysis was repeated for samples from that tissue.
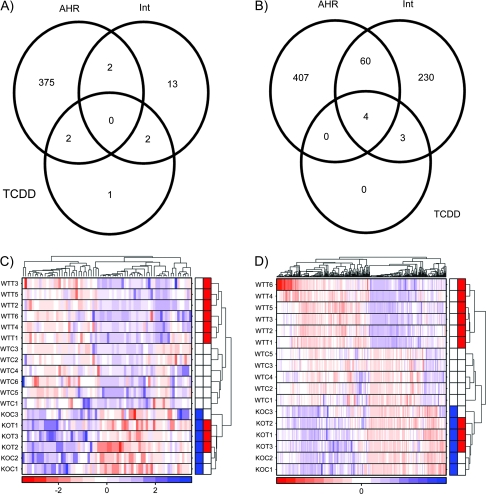
In the kidney (Fig. 2A) only five genes were altered by TCDD exposure in Ahr−/− mice, and the magnitudes of change were modest, ranging from 0.52- to 2.5-fold. Similarly, only seven genes are affected by TCDD in an AHR-independent manner in the liver (Fig. 2). Again, these changes are modest in magnitude, ranging from 0.52- to 2.22-fold. Thus a functional AHR is required for almost all TCDD-mediated alterations of transcript levels in mouse kidney and liver.
FIG. 2.
Effects of AHR−/− on transcriptional response to TCDD in kidney and liver. Following preprocessing of the microarray data, general-linear modeling was performed separately for the kidney (A) and liver (B) data. In both cases few genes were altered by TCDD in the absence of functional AHR protein whereas large numbers of genes were responsive to Ahr genotype even in the absence of TCDD exposure. Interestingly, whereas only 17 genes were TCDD responsive in an AHR-dependent fashion in kidney, fully 297 (17-fold more) were altered in liver. To visualize interanimal variability we employed unsupervised machine-learning separately on kidney (C) and liver (D); with few exceptions interanimal variability is small, and in all cases is smaller than intergroup variability. The color bar in the heat maps represents Ahr genotype (blue, AHR−/−; white, AHR+/+) and TCDD exposure (red, TCDD exposed; white, vehicle treated).
The two tissues were also similar in the number of genes exhibiting basal differences in mRNA levels between Ahr−/− and Ahr+/+ animals, even in the absence of TCDD exposure. In the kidney 379 genes exhibited AHR-dependent, TCDD-independent changes in mRNA levels—similar to the 471 genes altered in the liver.
Where the tissues differed significantly was the response to TCDD exposure. TCDD altered the mRNA levels of far fewer genes in mouse kidney than in mouse liver. Only 17 genes exhibited AHR-dependent mRNA alterations in response to TCDD exposure in mouse kidney (Table 1). By contrast 297 genes were altered in liver of these animals, and fully 22% (64/297) were also basally altered by Ahr genotype. A listing of all genes and their alterations in either tissue is in Supplementary Table 1. We confirmed that these conclusions were independent of our statistical threshold of padjusted < 0.05 with a threshold-analysis (Supplementary Fig. 1). To validate these findings, we employed real-time PCR. Five genes displaying AHR-dependent responses to TCDD and six genes responsive to Ahr status alone were tested. Nine of these eleven genes were validated (Supplementary Table 2).
TABLE 1.
mRNAs Altered by TCDD in Kidney and Liver
| Expression data |
||||||
| Entrez gene ID | Symbol | M (liver) | M (kidney) | p (liver) | p (kidney) | Name |
| 13076 | Cyp1a1 | 7.78 | 11.33 | 2.22 × 10−2 | 3.40 × 10−10 | Cytochrome P450, family 1, subfamily a, polypeptide 1 |
| 192113 | Atp12a | 0.00 | 3.05 | 1.00 × 100 | 4.67 × 10−2 | ATPase, H+/K+ transporting, nongastric, alpha polypeptide |
| 234593 | Ndrg4 | 0.00 | 2.12 | 1.00 × 100 | 3.63 × 10−4 | N-myc downstream regulated gene 4 |
| 56794 | Hacl1 | 0.75 | 1.96 | 7.66 × 10−2 | 5.03 × 10−5 | 2-Hydroxyacyl-CoA lyase 1 |
| 99929 | Tiparp | 5.98 | 1.25 | 1.14 × 10−7 | 3.63 × 10−4 | TCDD-inducible poly(ADP-ribose) polymerase |
| 76884 | Cyfip2 | 4.23 | 1.04 | 1.14 × 10−7 | 1.10 × 10−3 | Cytoplasmic FMR1 interacting protein 2 |
| 22217 | Usp12 | −0.02 | 0.88 | 6.02 × 10−1 | 7.41 × 10−3 | Ubiquitin specific peptidase 12 |
| 53415 | Htatip2 | 3.01 | 0.64 | 4.42 × 10−6 | 1.04 × 10−2 | HIV-1 tat interactive protein 2, homolog (human) |
| 18024 | Nfe2l2 | 1.99 | 0.55 | 4.87 × 10−5 | 3.46 × 10−2 | Nuclear factor, erythroid derived 2, like 2 |
| 73353 | Actrt2 | 0.01 | −0.03 | 1.00 × 100 | 1.95 × 10−2 | Actin-related protein T2 |
| 269704 | Zfp664 | 0.00 | −0.18 | 1.00 × 100 | 8.43 × 10−3 | Zinc finger protein 664 |
| 12484 | Cd24a | −0.19 | −0.37 | 8.46 × 10−1 | 3.68 × 10−3 | CD24a antigen |
| 12177 | Bnip3l | −0.36 | −0.38 | 3.04 × 10−1 | 3.46 × 10−2 | BCL2/adenovirus E1B interacting protein 3-like |
| 13645 | Egf | 0.00 | −0.40 | 1.00 × 100 | 1.07 × 10−2 | Epidermal growth factor |
| 20377 | Sfrp1 | 0.00 | −0.44 | 1.00 × 100 | 7.64 × 10−3 | Secreted frizzled-related protein 1 |
| 12192 | Zfp36l1 | −0.23 | −0.65 | 1.00 × 100 | 4.67 × 10−2 | Zinc finger protein 36, C3H type-like 1 |
| 13370 | Dio1 | −0.55 | −1.36 | 6.89 × 10−1 | 1.95 × 10−2 | Deiodinase, iodothyronine, type I |
Note. A listing of 17 genes whose mRNA abundances was altered by TCDD exposure in an AHR-dependent manner in kidney. The Entrez Gene ID is a stable gene identifier. The M values give the fold-changes of differential abundance in log2 space. For example, the M (kidney) of 3.05 for Atp12a indicates a 8.3-fold increase in abundance in Ahr+/+ animals exposed to TCDD. p Values have been subjected to a false-discovery rate adjustment for multiple testing.
Taken together these data indicate that Ahr genotype substantially affects the transcriptomes of liver and kidney, but that adult kidney is largely unaffected by TCDD exposure, at least at this time point and dose. To confirm this observation we took an unbiased approach and employed unsupervised machine learning (clustering) to identify the strongest trends in our data (Boutros and Okey, 2005). We selected the genes with the highest variance across samples and employed divisive hierarchical clustering. In both kidney (Fig. 2C) and liver (Fig. 2D) three clusters are observed. The first cluster (at the bottom of each heatmap with blue labels) contains all six Ahr−/− animals, with separation between TCDD-treated animals (with blue and red labels). The second cluster (in the centre of each heatmap) contains the wild-type, vehicle-treated animals (white labels). The final cluster (at the top of each heatmap) contains the TCDD-treated Ahr+/+ animals.
Comparison of Kidney and Liver
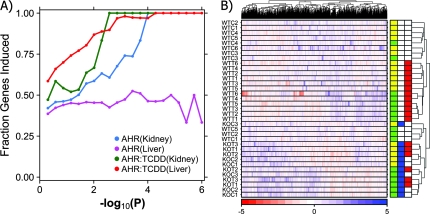
Previous studies by ourselves and others have demonstrated that the majority of TCDD-induced alterations in hepatic mRNA levels are inductive (Boutros et al., 2008; Boverhof et al., 2006; Fletcher et al., 2005). By contrast we previously showed that the majority of Ahr-associated changes in hepatic mRNA levels were repressive (Tijet et al., 2006). We studied the fraction of mRNA alterations that were inductive in each tissue (Fig. 3A). Intriguingly, our previous finding that Ahr-associated changes in hepatic mRNA abundances tend to be repressive is the outlier (purple curve), as the majority of TCDD-induced alterations in both tissues are inductive, as are the majority of genotype-dependent changes in kidney. Thus, although Ahr status alters mRNA abundances of large numbers of genes in both tissues, these alterations are different in character.
FIG. 3.
Characteristics of transcriptional response in kidney and liver. Following microarray preprocessing and general-linear modeling, we explored transcriptome-wide characteristics of genes altered in kidney and liver. Because very few genes displayed AHR-independent responses to TCDD these were not included in our analyses. First, we looked at the fraction of genes that were induced (A). Genes that were responsive to TCDD in either liver or kidney, as well as those altered by Ahr genotype in kidney, are much more likely to be inductive than repressive. By contrast genes responsive to Ahr genotype in liver were balanced slightly enriched for repressive responses. Next, to study intertissue responses to TCDD, we performed a coclustering analysis (B). The strongest trend was Ahr genotype, with both TCDD exposure and tissue having effects of similar magnitude. The color bar on the right side of the heatmap indicates tissue (yellow, kidney; green, liver), Ahr genotype (blue, AHR−/−; white, AHR+/+), and TCDD exposure (red, TCDD treated; white, vehicle treated).
To compare the trends between kidney and liver in an unbiased manner we again applied unsupervised machine-learning. Clustering of data from diverse tissues or datasets is a challenging problem because technical factors can obviate other trends. To solve this problem we applied our recently developed coclustering method (Boutros et al., 2008) to integrate the liver and kidney datasets (Fig. 3B). Three large clusters are evident. The first cluster (at the bottom of the heatmap) contains 15 samples, including all Ahr−/− animals (blue labels), which are subdivided by tissue (liver in green, kidney in yellow). Three wild-type (Ahr+/+) animals are also in this cluster. A second cluster, just above the first, comprises all TCDD-exposed animals, again subdivided by tissue. The final cluster (at the top of the heatmap) includes the remaining wild-type liver and kidney samples. Thus the strongest signal in the dataset is Ahr status: TCDD exposure and tissue play lesser roles. This suggests substantial overlap in genes affected by Ahr status in the two tissues.
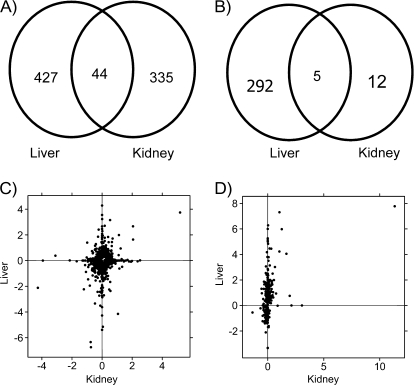
To explore this hypothesis we constructed Venn diagrams of genes displaying altered mRNA abundances based on Ahr status (Fig. 4A). In accord with the unsupervised machine-learning results, approximately 10% of the genes altered in either tissue are altered in both (9.3% in kidney, 11.6% in liver). This is far more than expected by chance alone (p < 10−64). A similar analysis of dioxin-responsive genes (Fig. 4B) highlights the paucity of dioxin-responsive genes in the kidney. Five genes are common to both tissues, a significant enrichment over chance expectations (p = 3.99 × 10−18).
FIG. 4.
Comparison of transcriptional response in kidney and liver. Following preprocessing and general-linear modeling of the microarray data, we compared the specific genes altered by Ahr genotype (A) and TCDD exposure (B) in liver and kidney. Large numbers of genes were altered by Ahr genotype in each tissue, and a significant fraction (p < 10−64) was common to both (A). By contrast, only five few genes were TCDD responsive in both tissues, which were again more than expected by chance (p = 3.99 × 10−18). To determine if these differences were related to genes just below our selected thresholds we plotted the fold-changes in liver against those in kidney for both genotype-responsive (C) and TCDD-responsive (D) genes. Although a number of genes lie on the diagonal in (C) many are along the axes indicating strong evidence of tissue specificity. Similarly, the TCDD-responsive genes are largely tissue-specific. The outlier in the top right of (D) with high fold changes in both liver and kidney is CYP1A1.
A subset of the 44 genes altered by Ahr genotype in both kidney and liver is shown in Table 2. The magnitudes of change vary dramatically, both within and between tissues. For example, metallothionein 1 mRNA levels are reduced by 99% in liver, but only by 40% in kidney. Some genes are altered in divergent directions: NADPH oxidase 4 (Nox4) and retinol saturase (Retsat) are induced in liver but repressed in kidney.
TABLE 2.
mRNAs Altered by Ahr Genotype
| Expression data |
||||||
| Entrez gene ID | Symbol | M (liver) | M (kidney) | p (liver) | p (kidney) | Name |
| 104923 | Adi1 | 3.74 | 5.18 | 3.52 × 10−4 | 1.70 × 10−3 | Acireductone dioxygenase 1 |
| 13370 | Dio1 | 1.63 | 2.06 | 7.28 × 10−4 | 1.92 × 10−5 | Deiodinase, iodothyronine, type I |
| 105892 | 9030619P08Rik | 0.42 | 1.10 | 1.39 × 10−2 | 3.17 × 10−3 | RIKEN cDNA 9030619P08 gene |
| 71670 | Acy3 | 0.79 | 0.62 | 2.61 × 10−3 | 2.28 × 10−3 | Aspartoacylase (aminoacylase) 3 |
| 108114 | Slc22a7 | 3.15 | 0.53 | 7.93 × 10−6 | 9.38 × 10−3 | Solute carrier family 22 (organic anion transporter), member 7 |
| 70337 | Iyd | −0.33 | 0.44 | 1.80 × 10−2 | 2.82 × 10−3 | Iodotyrosine deiodinase |
| 13476 | Reep5 | 0.57 | 0.38 | 8.55 × 10−3 | 8.45 × 10−3 | Receptor accessory protein 5 |
| 68347 | 0610011F06Rik | 0.49 | 0.37 | 3.76 × 10−3 | 3.17 × 10−3 | RIKEN cDNA 0610011F06 gene |
| 19703 | Renbp | −0.40 | −0.43 | 3.31 × 10−2 | 3.52 × 10−3 | Renin binding protein |
| 50490 | Nox4 | 0.90 | −0.44 | 2.22 × 10−2 | 8.40 × 10−3 | NADPH oxidase 4 |
| 15211 | Hexa | −0.60 | −0.46 | 2.84 × 10−4 | 1.70 × 10−3 | Hexosaminidase A |
| 109900 | Asl | −0.76 | −0.56 | 6.85 × 10−3 | 3.46 × 10−4 | Argininosuccinate lyase |
| 67442 | Retsat | 0.62 | −0.60 | 4.00 × 10−2 | 4.81 × 10−3 | Retinol saturase (all trans retinol 13,14 reductase) |
| 14873 | Gsto1 | −0.75 | −0.66 | 3.52 × 10−4 | 1.70 × 10−3 | Glutathione S-transferase omega 1 |
| 17748 | Mt1 | −6.74 | −0.74 | 3.83 × 10−6 | 1.70 × 10−3 | Metallothionein 1 |
Note. A listing of a subset of genes whose mRNA abundances were altered by Ahr genotype in both kidney and liver. The Entrez Gene ID is a stable gene identifier. The M values give the fold-changes of differential abundance in log2 space. For example, the M (Liver) of 3.74 for Adi1 indicates a 13.4-fold increase in abundance in the liver of animals with a wild-type Ahr relative to Ahr−/− animals. p Values have been subjected to a false-discovery rate adjustment for multiple testing.
The 17 genes whose mRNA renal abundances were altered by TCDD exposure in an Ahr-dependent manner are given in Table 1. The five altered in both liver and kidney include several well-characterized members of the AHR gene battery (Cyp1a1, Tiparp, Nfe2l2) along with two genes not well-established as AHR targets: cytoplasmic FMR1 interacting protein 2 (Cyfip2) and HIV-1 tat interactive protein 2 (Htatip2). Interestingly, the three AHR gene battery members are also altered in rat liver, whereas Cyfip2 and Htatip2 are not (Boutros et al., 2008). Htatip2 is, however, altered in the livers of mice with a constitutively active AHR (Moennikes et al., 2004). These results demonstrate that species-specific but tissue-independent AHR targets exist.
We then wondered if genes that were altered by either Ahr status or TCDD exposure in one tissue were predisposed to show weak, but statistically insignificant changes in the other. This would represent “leaky control,” where AHR regulation was not completely tissue specific. To address this hypothesis we generated scatter plots of the fold changes (in log2 space) of liver and kidney both for genes altered by Ahr status (Fig. 4C) and by TCDD exposure (Fig. 4D). In both cases we observe a “cloud” of points near the origin, with many genes found directly on either the x-axis (indicating kidney-specific changes) or the y-axis (indicating liver-specific changes). Because only a few genes lie on the diagonal, we can discount the idea of “leaky control” and conclude that the majority of genes have strongly tissue-specific effects. The outlier in the top right corner of Figure 4C is acireductone dioxygenase 1 (Adi1); the outlier in the top right corner of Figure 4D is Cyp1a1.
Pathways Responsive to Ahr Genotype and TCDD
Although 44 genes respond to Ahr genotype in both liver and kidney (Fig. 4A) and five respond to TCDD in both tissues (Fig. 4B), far more genes display tissue-specific responses. There are 762 tissue-specific responses to Ahr genotype and 309 to TCDD exposure. However it remains possible that the AHR has similar functions in each tissue, but executes them through regulation of different genes. To test this hypothesis, as well as to determine the specific pathways perturbed by Ahr genotype and TCDD exposure, we performed a GO enrichment analysis (Zeeberg et al., 2003). We divided the dataset into sets of genes showing tissue-specific and tissue-independent responses to AHR genotype and TCDD exposure (i.e., the six components of the Venn diagrams in Figs. 4A and 4B). We performed GO analysis separately on each of these six gene lists.
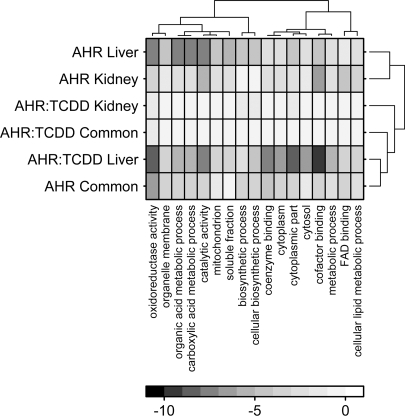
We focused on those GO terms showing robust alterations: those with a cumulative p value below 10−12 (Fig. 5). A total of 17 GO terms met this threshold, and these represent GO terms describing subcellular localizations (e.g., mitochondrial localization), molecular functions (e.g., oxidoreductase activity), and biological processes (e.g., cellular lipid metabolism). Intriguingly, the set of GO terms altered by Ahr genotype in kidney is quite similar to that in liver (Pearson's R = 0.364, p < 2.2 × 10−16). Similarly, the set of genes altered by TCDD exposure in liver is quite similar to those altered by Ahr genotype in both liver and kidney (Pearson's R = 0.467, p < 2.2 × 10−16). Taken together, these results indicate that the AHR can affect some of the same pathways in liver and kidney. In some cases it does so through tissue-specific alterations, whereas in other cases the same genes are affected in both tissues. Intriguingly those pathways dysregulated by the same genes in both tissues are also those most likely to be perturbed by TCDD, at least in liver. Complete GO results are in Supplementary Table 3.
FIG. 5.
Functional analysis of perturbed genes. GO enrichment analysis was performed on the sets of genes altered in response to Ahr genotype or TCDD exposure in kidney only, liver only, or both tissues. The log10(p values) were calculated for each GO term and the 17 terms with pcumulative < 10−12 are shown here. Unsupervised hierarchical clustering was used to determine relationships amongst GO terms and tissues. The color-bar indicates log10(p values), so dark squares indicate highly statistically significant enrichments.
Transcription Factors Involved in Tissue-Specific Responses
To understand the pathways involved in genotype- and TCDD-dependent changes in mRNA abundance we employed computational techniques to assess motifs of TFBS. We first focused on three TFBSs closely-associated with AHR activity: the two aryl hydrocarbon response elements, AHRE-I and AHRE-II, along with the ARE. The regulatory region (−3000 to +3000) of each gene whose mRNA levels were altered by Ahr genotype or TCDD exposure was extracted and searched for each TFBS, and the evolutionary conservation of all motifs was determined using PhastCons scores (Boutros et al., 2004; Siepel and Haussler, 2004). PhastCons scores range from 0 (no conservation across 30 vertebrate species) to 1 (perfect conservation across 30 vertebrate species). This analysis was repeated for genes altered by Ahr genotype in kidney only, liver only, or both tissues and for genes altered by TCDD exposure in kidney only, liver only, or both tissues.
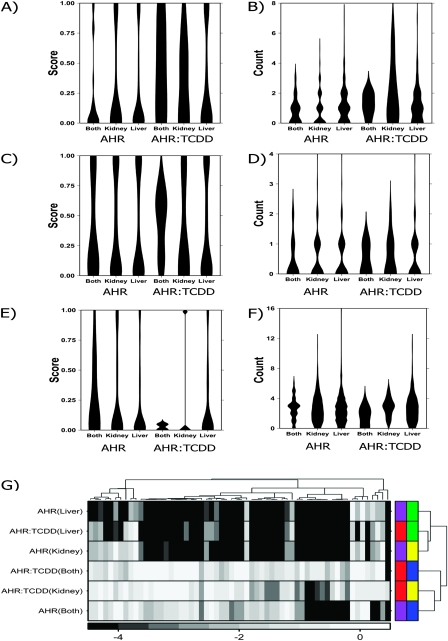
The resulting six lists were visualized using violin plots for AHRE-I (Figs. 6A and 6B), AHRE-II (Figs. 6C and 6D), and ARE (Figs. 6E and 6F). Violin plots are a compact method of representing the distribution of a data set. Each column represents a specific condition, and the thickness of a column represents the fraction of data points with that value (i.e., the density of the distribution). Thus violin plots allow the simultaneous visualization of multiple distributions. For example, Figure 6A shows that AHRE-I motifs present in genes responsive to TCDD in both liver and kidney are more likely to be evolutionarily conserved than those in the other gene lists. Similarly, Figure 6B shows that genes responsive to TCDD in kidney contain more AHRE-I motifs than those in the other gene lists. Complete data on the number and conservation of motifs for each gene is in Supplementary Table 1.
FIG. 6.
Analysis of TFBS. TFBSs were analyzed in regulatory sequence from −3000 to +3000 bp relative to the transcriptional start-site of each gene altered in one or both tissues by either Ahr genotype (AHR) or TCDD exposure (AHR:TCDD). Three TFBS motifs known to be associated with AHR activity were considered: AHRE-I (A and B), AHRE-II (C and D), and ARE (E and F). For each gene, the phylogenetic conservation of motifs was calculated on a scale of zero (low conservation) to one (high conservation) (parts A, C, and E). Additionally the actual number of motifs was calculated (parts B, D, and F). The visualizations used for parts A–F are called “violin plots”. Each column represents a specific gene list, and the thickness and shape of the column represents the distribution of values for that gene list. A library-based TFBS-enrichment analysis was performed on the same sequence and the log10|p| from this analysis subjected to unsupervised clustering (G). The color bar indicates log10|p| values. The permutation test employed had a minimum resolution of 10−4, hence the large number of ties at this level. The color bars on the right side reflect tissue of origin (liver, green; kidney, yellow; both tissues, blue) and contrast (AHR genotype, purple; TCDD-induced changes, red).
This analysis raises several intriguing points. Many genes whose mRNA abundances are altered by TCDD display AHRE-I motifs (Fig. 6B) which are often conserved, especially in genes altered in both tissues (Fig. 6A). Genes altered in both tissues also tend to have more (Fig. 6D) and better-conserved (Fig. 6C) AHRE-II motifs. This suggests that known tissue-independent responses to TCDD may occur through well-characterized mechanisms of AHR activity. Genes altered by Ahr genotype, on the other hand, are slightly enriched for AREs (Figs. 6E and 6F), whereas TCDD-dependent responses are not.
These results suggest that other transcription factors may collaborate with the AHR in a combinatorial fashion to regulate tissue-specific mRNA regulation. To assess this effect we performed a library-based TFBS analysis, searching the regulatory regions of each gene list for 130 known TFBS motifs. Only motifs enriched (p < 0.05) in three separate statistical tests were considered. In total 60 TFBSs met this criterion in one or more gene lists (Supplementary Table 4). Unsupervised clustering was used on these motifs to determine which gene lists shared the most regulatory structure (Fig. 6G). The six lists divided into two clusters. The first contained genes responsive to Ahr genotype in a tissue-specific manner (i.e., liver only and kidney only) and genes responsive to TCDD in the liver. The second cluster contained tissue-independent responses to Ahr genotype or TCDD activity and the genes responsive to TCDD in the kidney alone. This division further supports the concept that tissue-specific and tissue-independent responses occur through distinct mechanisms.
DISCUSSION
Dioxin-induced toxicities affect many tissues (McConnell et al., 1978; Pohjanvirta and Tuomisto, 1994; Pohjanvirta et al, 1989). Although it is becoming well-established that the vast majority of these toxicities are mediated through the AHR, the pathways leading from AHR activation to most specific toxicities remain unknown (Okey, 2007). There is also accumulating evidence that the AHR plays a critical role in normal development (McMillan and Bradfield, 2007), perhaps through activation by endogenous ligands (Bittinger et al., 2003; Nguyen and Bradfield, 2008; Song et al., 2002).
Because the AHR is a transcription factor, one way to assess its activity is to profile the transcriptional response to TCDD exposure in wild-type (Ahr+/+) or AHR-null (Ahr−/−) mice. When we studied the response of adult liver to TCDD exposure in this way three trends emerged (Tijet et al., 2006). First, the transcriptional effects of TCDD required the presence of functional AHR protein. That is, Ahr−/− mice are not only phenotypically refractory to TCDD exposure, but also show virtually no cellular response. Second, large numbers of genes in specific functional pathways are perturbed by dioxin exposure in Ahr+/+ mice. Third, surprisingly large numbers of genes are perturbed by genetic ablation of the Ahr, even in absence of exogenous ligand. These genes lie in specific functional pathways and are associated with specific TFBS, suggesting an important role for AHR activity in basal hepatic physiology.
Here, we replicate our hepatic study in kidney, performing transcriptional profiling of adult Ahr−/− and Ahr+/+ mice with or without exposure to TCDD. We extensively compared the effects of dioxin exposure and Ahr genotype in kidney and liver. We took numerous steps to ensure that this comparison could be made robustly and with minimal noise. First, tissues were excised from the same animals at the same time, thereby minimizing interanimal or interbatch variation. Second, RNA preparations were performed in an identical manner. Third, microarray hybridizations were performed using identical platforms and protocols. Fourth, computational and statistical analyses were performed identically, using the same software versions and tests for each dataset. We note that we selected a high dose, equivalent to roughly three times the LD50 and an early time point so that we could capture acute and early effects of dioxin exposure.
The renal and hepatic effects of dioxin exposure and Ahr genotype differ substantially. Although large numbers of genes are affected by each factor in the liver, kidney was mostly refractory to dioxin exposure. The exceptions (Table 1, Fig. 4D) include well-characterized AHR-responsive genes, such as Cyp1a1 and Tiparp. These data emphasize the tissue-specific regulatory effects of the AHR. Indeed our analysis of TFBS identified motifs associated with AHR activity in a tissue-specific manner.
Surprisingly, the kidney transcriptome was substantially remodeled in the absence of a functional AHR. This might indicate a developmental role for the AHR in this tissue, or may indicate a role for the AHR in adult renal physiology. There are at least two known renal phenotypes in Ahr−/− mice. First, kidney weight is higher in Ahr−/− males than in wild-type males, at least at three months of age (Lin et al., 2001). Second, neonatal Ahr−/− mice exhibit significant vascular abnormalities in the kidney (Lahvis et al., 2000; McMillan and Bradfield, 2007). The underlying mechanism has not been investigated but could contribute to the age-related hypertension observed in adult Ahr−/− mice (Lund et al., 2008).
Several studies have considered the effects of Ahr activation on fetal mouse kidney (Johnson et al., 2004). In such studies, as well as in our own work (Choi et al., 2006) there is substantial evidence of CYP1B1 induction by AHR ligands. Surprisingly, CYP1B1 induction has not been observed in adult mouse kidney (Kopf et al., 2008; Shimada et al., 2003), although it has in adult rat kidney (Badawi et al., 2000). We confirm these results by observing no upregulation of CYP1B1 mRNA in adult mouse kidney. However we do not observe constitutive overexpression of CYP1B1 mRNA in Ahr−/− kidneys, which has been reported by others (Shimada et al., 2003). Taken together, these results suggest substantial differences in the renal transcriptional response to TCDD between adult and fetal mice, as well as mice and rats.
Although the adult liver is susceptible to acute TCDD-induced toxicity, the adult kidney is not. To the best of our knowledge, the lack of impact of TCDD on adult mouse kidney has not specifically studied in detail (Birnbaum and Tuomisto, 2000). However a very recent paper (Lu et al., 2009) has studied the response of rat kidney to a cumulative dose of 120 μg/kg TCDD in a 12-day experiment. The authors detected several signs of nephrotoxicity, including increased serum creatinine and blood urea nitrogen, histopathological changes, and increased renal oxidative stress. It is not clear, however, if these are primary effects of TCDD as these animals exhibited reduced activity, a 27% decline in food intake and a 28% decline in body-weight (feed-restricted controls were not included). On balance, then, it appears that adult kidney is not acutely responsive to TCDD, perhaps in part because TCDD is sequestered in liver and does not distribute to the kidney (Santostefano et al., 1996). It is known, however, that chronic low doses result in hypertension which may be due, in part, to effects on kidney (Kopf et al., 2008). In this context, the small number of TCDD-induced changes observed in kidney is reasonable. We hypothesize that significantly more genes will be altered by TCDD in fetal kidney.
Our conclusions about tissue specificity must be taken with at least one caveat, however. A tissue is not a homogeneous entity, but rather is a complex mixture of different cell-types. About 80% of liver volume is comprised of hepatocytes, whereas endothelial, Kupffer, stellate, and intrahepatic lymphocytes make up the remaining 20% (Lieberman et al., 2009). By contrast, kidney consists of distinct structural and functional components which, together with the supporting vasculature, comprise a large number of cell types. At least twelve cell types have been identified in the nephron, ranging in function from maintenance of the filtration barrier (glomerular epithelium) to selective reabsorption of glucose and amino acids (proximal tubular epithelium) (Gilbert, 2000; Rosenblum, 2008). A large transcriptional effect observed in a relatively rare cell-type might be difficult to detect in our experiments. Further complications would arise if different cell-types have divergent responses, as these may cancel one another out. These factors might inflate our false-negative rate and our results represent a lower-bound on the number of genes altered by dioxin exposure and Ahr genotype.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
Canadian Institutes for Health Research (grant no. MOP-57903 to P.C.B. and MOP-53178 to P.A.H.); and the Academy of Finland (grant no. 123345) to R.P.
Acknowledgments
The authors thank Dr Allan Okey for insightful commentary at all stages of this project.
References
- Abbott BD, Birnbaum LS. Effects of TCDD on embryonic ureteric epithelial EGF receptor expression and cell proliferation. Teratology. 1990;41:71–84. doi: 10.1002/tera.1420410108. [DOI] [PubMed] [Google Scholar]
- Abbott BD, Birnbaum LS, Pratt RM. TCDD-induced hyperplasia of the ureteral epithelium produces hydronephrosis in murine fetuses. Teratology. 1987;35:329–334. doi: 10.1002/tera.1420350307. [DOI] [PubMed] [Google Scholar]
- Badawi AF, Cavalieri EL, Rogan EG. Effect of chlorinated hydrocarbons on expression of cytochrome P450 1A1, 1A2 and 1B1 and 2- and 4-hydroxylation of 17beta-estradiol in female Sprague-Dawley rats. Carcinogenesis. 2000;21:1593–1599. [PubMed] [Google Scholar]
- Beischlag TV, Wang S, Rose DW, Torchia J, Reisz-Porszasz S, Muhammad K, Nelson WE, Probst MR, Rosenfeld MG, Hankinson O. Recruitment of the NCoA/SRC-1/p160 family of transcriptional coactivators by the aryl hydrocarbon receptor/aryl hydrocarbon receptor nuclear translocator complex. Mol. Cell. Biol. 2002;22:4319–4333. doi: 10.1128/MCB.22.12.4319-4333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum LS, Tuomisto J. Non-carcinogenic effects of TCDD in animals. Food Addit. Contam. 2000;17:275–288. doi: 10.1080/026520300283351. [DOI] [PubMed] [Google Scholar]
- Bittinger MA, Nguyen LP, Bradfield CA. Aspartate aminotransferase generates proagonists of the aryl hydrocarbon receptor. Mol. Pharmacol. 2003;64:550–556. doi: 10.1124/mol.64.3.550. [DOI] [PubMed] [Google Scholar]
- Boutros PC, Moffat ID, Franc MA, Tijet N, Tuomisto J, Pohjanvirta R, Okey AB. Dioxin-responsive AHRE-II gene battery: Identification by phylogenetic footprinting. Biochem. Biophys. Res. Commun. 2004;321:707–715. doi: 10.1016/j.bbrc.2004.06.177. [DOI] [PubMed] [Google Scholar]
- Boutros PC, Okey AB. Unsupervised pattern recognition: An introduction to the whys and wherefores of clustering microarray data. Brief Bioinform. 2005;6:331–343. doi: 10.1093/bib/6.4.331. [DOI] [PubMed] [Google Scholar]
- Boutros PC, Yan R, Moffat ID, Pohjanvirta R, Okey AB. Transcriptomic responses to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in liver: Comparison of rat and mouse. BMC Genomics. 2008;9:419. doi: 10.1186/1471-2164-9-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boverhof DR, Burgoon LD, Tashiro C, Sharratt B, Chittim B, Harkema JR, Mendrick DL, Zacharewski TR. Comparative toxicogenomic analysis of the hepatotoxic effects of TCDD in Sprague Dawley rats and C57BL/6 mice. Toxicol. Sci. 2006;94:398–416. doi: 10.1093/toxsci/kfl100. [DOI] [PubMed] [Google Scholar]
- Choi SS, Miller MA, Harper PA. In utero exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin induces amphiregulin gene expression in the developing mouse ureter. Toxicol. Sci. 2006;94:163–174. doi: 10.1093/toxsci/kfl090. [DOI] [PubMed] [Google Scholar]
- Couture LA, Harris MW, Birnbaum LS. Characterization of the peak period of sensitivity for the induction of hydronephrosis in C57BL/6N mice following exposure to 2,3,7, 8-tetrachlorodibenzo-p-dioxin. Fundam. Appl. Toxicol. 1990;15:142–150. doi: 10.1016/0272-0590(90)90171-f. [DOI] [PubMed] [Google Scholar]
- Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron B, Tibshirani R. Empirical Bayes methods and false discovery rates for microarrays. Genet. Epidemiol. 2002;23:70–86. doi: 10.1002/gepi.1124. [DOI] [PubMed] [Google Scholar]
- Fletcher N, Wahlstrom D, Lundberg R, Nilsson CB, Nilsson KC, Stockling K, Hellmold H, Hakansson H. 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) alters the mRNA expression of critical genes associated with cholesterol metabolism, bile acid biosynthesis, and bile transport in rat liver: A microarray study. Toxicol. Appl. Pharmacol. 2005;207:1–24. doi: 10.1016/j.taap.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Franc MA, Moffat ID, Boutros PC, Tuomisto JT, Tuomisto J, Pohjanvirta R, Okey AB. Patterns of dioxin-altered mRNA expression in livers of dioxin-sensitive versus dioxin-resistant rats. Arch. Toxicol. 2008;82:809–830. doi: 10.1007/s00204-008-0303-0. [DOI] [PubMed] [Google Scholar]
- Frith MC, Fu Y, Yu L, Chen JF, Hansen U, Weng Z. Detection of functional DNA motifs via statistical over-representation. Nucleic Acids Res. 2004;32:1372–1381. doi: 10.1093/nar/gkh299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frueh FW, Hayashibara KC, Brown PO, Whitlock JP., Jr Use of cDNA microarrays to analyze dioxin-induced changes in human liver gene expression. Toxicol. Lett. 2001;122:189–203. doi: 10.1016/s0378-4274(01)00364-2. [DOI] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA. affy—Analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- Gilbert SF. Developmental Biology. Sunderland, MA: Sinauer Associates; 2000. [Google Scholar]
- Hankinson O. The aryl hydrocarbon receptor complex. Annu. Rev. Pharmacol. Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- Hankinson O. Role of coactivators in transcriptional activation by the aryl hydrocarbon receptor. Arch. Biochem. Biophys. 2005;433:379–386. doi: 10.1016/j.abb.2004.09.031. [DOI] [PubMed] [Google Scholar]
- Huuskonen H, Unkila M, Pohjanvirta R, Tuomisto J. Developmental toxicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in the most TCDD-resistant and -susceptible rat strains. Toxicol. Appl. Pharmacol. 1994;124:174–180. doi: 10.1006/taap.1994.1021. [DOI] [PubMed] [Google Scholar]
- Johnson CD, Balagurunathan Y, Tadesse MG, Falahatpisheh MH, Brun M, Walker MK, Dougherty ER, Ramos KS. Unraveling gene-gene interactions regulated by ligands of the aryl hydrocarbon receptor. Environ. Health Perspect. 2004;112:403–412. doi: 10.1289/ehp.6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, Roskin KM, Schwartz M, Sugnet CW, Thomas DJ, et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A, Sogawa K, Fujii-Kuriyama Y. Cooperative interaction between AhR.Arnt and Sp1 for the drug-inducible expression of CYP1A1 gene. J. Biol. Chem. 1996;271:12310–12316. doi: 10.1074/jbc.271.21.12310. [DOI] [PubMed] [Google Scholar]
- Kopf PG, Huwe JK, Walker MK. Hypertension, cardiac hypertrophy, and impaired vascular relaxation induced by 2,3,7,8-tetrachlorodibenzo-p-dioxin are associated with increased superoxide. Cardiovasc. Toxicol. 2008;8:181–193. doi: 10.1007/s12012-008-9027-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahvis GP, Lindell SL, Thomas RS, McCuskey RS, Murphy C, Glover E, Bentz M, Southard J, Bradfield CA. Portosystemic shunting and persistent fetal vascular structures in aryl hydrocarbon receptor-deficient mice. Proc. Natl. Acad. Sci. U. S. A. 2000;97:10442–10447. doi: 10.1073/pnas.190256997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman M, Marks AD, Smith CM. Marks’ Basic Medical Biochemistry: A Clinical Approach. Philadelphia, PA: Wolters Kluwer/Lippincott Williams & Wilkins Health; 2009. [Google Scholar]
- Lin BC, Nguyen LP, Walisser JA, Bradfield CA. A hypomorphic allele of aryl hydrocarbon receptor-associated protein-9 produces a phenocopy of the AHR-null mouse. Mol. Pharmacol. 2008;74:1367–1371. doi: 10.1124/mol.108.047068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin TM, Ko K, Moore RW, Buchanan DL, Cooke PS, Peterson RE. Role of the aryl hydrocarbon receptor in the development of control and 2,3,7,8-tetrachlorodibenzo-p-dioxin-exposed male mice. J. Toxicol. Environ. Health A. 2001;64:327–342. doi: 10.1080/152873901316981312. [DOI] [PubMed] [Google Scholar]
- Lu CF, Wang YM, Peng SQ, Zou LB, Tan DH, Liu G, Fu Z, Wang QX, Zhao J. Combined effects of repeated administration of 2,3,7,8-tetrachlorodibenzo-p-dioxin and polychlorinated biphenyls on kidneys of male rats. Arch. Environ. Contam. Toxicol. 2009 doi: 10.1007/s00244-009-9323-x. (Forthcoming) [DOI] [PubMed] [Google Scholar]
- Lund AK, Agbor LN, Zhang N, Baker A, Zhao H, Fink GD, Kanagy NL, Walker MK. Loss of the aryl hydrocarbon receptor induces hypoxemia, endothelin-1, and systemic hypertension at modest altitude. Hypertension. 2008;51:803–809. doi: 10.1161/HYPERTENSIONAHA.107.100586. [DOI] [PubMed] [Google Scholar]
- Matsushita N, Sogawa K, Ema M, Yoshida A, Fujii-Kuriyama Y. A factor binding to the xenobiotic responsive element (XRE) of P-4501A1 gene consists of at least two helix-loop-helix proteins, Ah receptor and Arnt. J. Biol. Chem. 1993;268:21002–21006. [PubMed] [Google Scholar]
- McConnell EE, Moore JA, Haseman JK, Harris MW. The comparative toxicity of chlorinated dibenzo-p-dioxins in mice and guinea pigs. Toxicol. Appl. Pharmacol. 1978;44:335–356. doi: 10.1016/0041-008x(78)90195-3. [DOI] [PubMed] [Google Scholar]
- McMillan BJ, Bradfield CA. The aryl hydrocarbon receptor sans xenobiotics: Endogenous function in genetic model systems. Mol. Pharmacol. 2007;72:487–498. doi: 10.1124/mol.107.037259. [DOI] [PubMed] [Google Scholar]
- Moennikes O, Loeppen S, Buchmann A, Andersson P, Ittrich C, Poellinger L, Schwarz M. A constitutively active dioxin/aryl hydrocarbon receptor promotes hepatocarcinogenesis in mice. Cancer Res. 2004;64:4707–4710. doi: 10.1158/0008-5472.CAN-03-0875. [DOI] [PubMed] [Google Scholar]
- Moffat ID, Boutros PC, Celius T, Linden J, Pohjanvirta R, Okey AB. microRNAs in adult rodent liver are refractory to dioxin treatment. Toxicol. Sci. 2007;99:470–487. doi: 10.1093/toxsci/kfm189. [DOI] [PubMed] [Google Scholar]
- Muller PY, Janovjak H, Miserez AR, Dobbie Z. Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques. 2002;32:1372–1374. 1376, 1378–1379. [PubMed] [Google Scholar]
- Nguyen LP, Bradfield CA. The search for endogenous activators of the aryl hydrocarbon receptor. Chem. Res. Toxicol. 2008;21:102–116. doi: 10.1021/tx7001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TA, Hoivik D, Lee JE, Safe S. Interactions of nuclear receptor coactivator/corepressor proteins with the aryl hydrocarbon receptor complex. Arch. Biochem. Biophys. 1999;367:250–257. doi: 10.1006/abbi.1999.1282. [DOI] [PubMed] [Google Scholar]
- Ohtake F, Takeyama K, Matsumoto T, Kitagawa H, Yamamoto Y, Nohara K, Tohyama C, Krust A, Mimura J, Chambon P, et al. Modulation of oestrogen receptor signalling by association with the activated dioxin receptor. Nature. 2003;423:545–550. doi: 10.1038/nature01606. [DOI] [PubMed] [Google Scholar]
- Okey AB. An aryl hydrocarbon receptor odyssey to the shores of toxicology: the Deichmann Lecture, International Congress of Toxicology-XI. Toxicol. Sci. 2007;98:5–38. doi: 10.1093/toxsci/kfm096. [DOI] [PubMed] [Google Scholar]
- Pohjanvirta R. Transgenic mouse lines expressing rat AH receptor variants—A new animal model for research on AH receptor function and dioxin toxicity mechanisms. Toxicol. Appl. Pharmacol. 2009;236:166–182. doi: 10.1016/j.taap.2009.01.005. [DOI] [PubMed] [Google Scholar]
- Pohjanvirta R, Kulju T, Morselt AF, Tuominen R, Juvonen R, Rozman K, Mannisto P, Collan Y, Sainio EL, Tuomisto J. Target tissue morphology and serum biochemistry following 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) exposure in a TCDD-susceptible and a TCDD-resistant rat strain. Fundam. Appl. Toxicol. 1989;12:698–712. doi: 10.1016/0272-0590(89)90002-x. [DOI] [PubMed] [Google Scholar]
- Pohjanvirta R, Tuomisto J. Short-term toxicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin in laboratory animals: Effects, mechanisms, and animal models. Pharmacol. Rev. 1994;46:483–549. [PubMed] [Google Scholar]
- Puga A, Maier A, Medvedovic M. The transcriptional signature of dioxin in human hepatoma HepG2 cells. Biochem. Pharmacol. 2000;60:1129–1142. doi: 10.1016/s0006-2952(00)00403-2. [DOI] [PubMed] [Google Scholar]
- Quintana FJ, Basso AS, Iglesias AH, Korn T, Farez MF, Bettelli E, Caccamo M, Oukka M, Weiner HL. Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature. 2008;453:65–71. doi: 10.1038/nature06880. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. 2008 R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0. Available at http://www.R-project.org. Accessed September 22, 2009. [Google Scholar]
- Rosenblum ND. Developmental biology of the human kidney. Semin. Fetal Neonatal Med. 2008;13:125–132. doi: 10.1016/j.siny.2007.10.005. [DOI] [PubMed] [Google Scholar]
- Rowlands JC, McEwan IJ, Gustafsson JA. Trans-activation by the human aryl hydrocarbon receptor and aryl hydrocarbon receptor nuclear translocator proteins: Direct interactions with basal transcription factors. Mol. Pharmacol. 1996;50:538–548. [PubMed] [Google Scholar]
- Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B. JASPAR: An open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32(Database issue):D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santostefano MJ, Johnson KL, Whisnant NA, Richardson VM, DeVito MJ, Diliberto JJ, Birnbaum LS. Subcellular localization of TCDD differs between the liver, lungs, and kidneys after acute and subchronic exposure: Species/dose comparisons and possible mechanism. Fundam. Appl. Toxicol. 1996;34:265–275. doi: 10.1006/faat.1996.0196. [DOI] [PubMed] [Google Scholar]
- Schmidt JV, Su GH, Reddy JK, Simon MC, Bradfield CA. Characterization of a murine Ahr null allele: Involvement of the Ah receptor in hepatic growth and development. Proc. Natl. Acad. Sci. U. S. A. 1996;93:6731–6736. doi: 10.1073/pnas.93.13.6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Sugie A, Shindo M, Nakajima T, Azuma E, Hashimoto M, Inoue K. Tissue-specific induction of cytochromes P450 1A1 and 1B1 by polycyclic aromatic hydrocarbons and polychlorinated biphenyls in engineered C57BL/6J mice of arylhydrocarbon receptor gene. Toxicol. Appl. Pharmacol. 2003;187:1–10. doi: 10.1016/s0041-008x(02)00035-2. [DOI] [PubMed] [Google Scholar]
- Siepel A, Haussler D. Combining phylogenetic and hidden Markov models in biosequence analysis. J. Comput. Biol. 2004;11:413–428. doi: 10.1089/1066527041410472. [DOI] [PubMed] [Google Scholar]
- Simon P. Q-Gene: Processing quantitative real-time RT-PCR data. Bioinformatics. 2003;19:1439–1440. doi: 10.1093/bioinformatics/btg157. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2003;3:1–26. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Sogawa K, Numayama-Tsuruta K, Takahashi T, Matsushita N, Miura C, Nikawa J, Gotoh O, Kikuchi Y, Fujii-Kuriyama Y. A novel induction mechanism of the rat CYP1A2 gene mediated by Ah receptor-Arnt heterodimer. Biochem. Biophys. Res. Commun. 2004;318:746–755. doi: 10.1016/j.bbrc.2004.04.090. [DOI] [PubMed] [Google Scholar]
- Song J, Clagett-Dame M, Peterson RE, Hahn ME, Westler WM, Sicinski RR, DeLuca HF. A ligand for the aryl hydrocarbon receptor isolated from lung. Proc. Natl. Acad. Sci. U. S. A. 2002;99:14694–14699. doi: 10.1073/pnas.232562899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijet N, Boutros PC, Moffat ID, Okey AB, Tuomisto J, Pohjanvirta R. Aryl hydrocarbon receptor regulates distinct dioxin-dependent and dioxin-independent gene batteries. Mol. Pharmacol. 2006;69:140–153. doi: 10.1124/mol.105.018705. [DOI] [PubMed] [Google Scholar]
- Veldhoen M, Hirota K, Westendorf AM, Buer J, Dumoutier L, Renauld JC, Stockinger B. The aryl hydrocarbon receptor links T(H)17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453:106–109. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- Walisser JA, Bunger MK, Glover E, Harstad EB, Bradfield CA. Patent ductus venosus and dioxin resistance in mice harboring a hypomorphic Arnt allele. J. Biol. Chem. 2004;279:16326–16331. doi: 10.1074/jbc.M400784200. [DOI] [PubMed] [Google Scholar]
- Wu Z, Irizarry RA, Gentleman R, Murillo FM, Spencer R. A model based background adjustment for oligonucleotide expression arrays. Johns Hopkins Univ. Dept. Biostat. Working Papers. 2004;1:1–26. [Google Scholar]
- Zeeberg BR, Feng W, Wang G, Wang MD, Fojo AT, Sunshine M, Narasimhan S, Kane DW, Reinhold WC, Lababidi S, et al. GoMiner: A resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003;4:R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.