Abstract
Salmonella enterica serovar Choleraesuis (Salmonella Choleraesuis) causes a lethal systemic infection (salmonellosis) in swine. Live attenuated Salmonella Choleraesuis vaccines are effective in preventing the disease, and isolates of Salmonella Choleraesuis with mutations in the cAMP-receptor protein (CRP) gene (Salmonella Choleraesuis ∆crp) are the most widely used, although the basis of the attenuation remains unclear. The objective of this study was to determine if the attenuated phenotype of Salmonella Choleraesuis ∆crp was due to alterations in susceptibility to gastrointestinal factors such as pH and bile salts, ability to colonize or invade the intestine, or cytotoxicity for macrophages. Compared with the parental strain, the survival rate of Salmonella Choleraesuis ∆crp at low pH or in the presence of bile salts was higher, while the ability of the mutant to invade intestinal epithelia was significantly decreased. In examining the role of CRP on the secretory function of the Salmonella pathogenicity island 1 (SPI-1) encoded type III secretion system (T3SS), it was shown that Salmonella Choleraesuis ∆crp was unable to secrete the SPI-1 T3SS effector proteins, SopB and SipB, which play a role in Salmonella intestinal invasiveness and macrophage cytotoxicity, respectively. In addition, caspase-1 dependent cytotoxicity for macrophages was significantly reduced in Salmonella Choleraesuis ∆crp. Collectively, this study demonstrates that the CRP affects the secretory function of SPI-1 T3SS and the resulting ability to invade the host intestinal epithelium, which is a critical element in the pathogenesis of Salmonella Choleraesuis.
Keywords: Salmonella enterica serovar Choleraesuis, cAMP-receptor protein, CRP, type III secretion system, attenuation
1. INTRODUCTION
Salmonella enterica serovar Choleraesuis (herein termed Salmonella Choleraesuis) is one of the prevalent serovars causing salmonellosis in swine. Clinically, pigs infected by Salmonella Choleraesuis develop not only gastroenteritis, but also lethal systemic manifestations including pneumonia, hepatitis, and meningitis [32]. Although Salmonella Choleraesuis is a host-adapted pathogen of pigs, studies have revealed this serovar may also cause systemic infections in humans [6, 7]; however, the relationship between isolates of Salmonella Choleraesuis from humans and swine remains to be elucidated [4].
Vaccines represent an effective strategy for the control of swine salmonellosis [32]. However, bacterins composed of inactivated cultures of Salmonella are less effective than live vaccines consisting of attenuated strains of Salmonella [15]. During the past decades, a variety of live attenuated Salmonella Choleraesuis mutants have been developed as vaccine candidates. These include strains having mutations in genes involved in purine synthesis (pur), thymidine synthesis (thy), the cAMP-receptor protein (crp), adenylate cyclase (cya), type III secretion system (T3SS) apparatus (ssaC, ssaJ and ssaV), Gifsy-1 prophage protein (gifsy-1), and D-galactonate transport protein (dgoT) [8, 19–21, 27]. Among these, Salmonella Choleraesuis strains with mutations in crp alone or in combination with other genes were shown to be greatly attenuated in virulence and to serve as effective vaccine candidates against swine salmonellosis [8, 20, 34].
In Escherichia coli, the cAMP-receptor protein (CRP) activates transcription of more than 100 genes and acts as a global gene regulator [22]. Upon coupling with the allosteric effector, cyclic AMP, dimeric CRP initiates transcription by binding to specific DNA sites and enhancing the binding of RNA polymerase holoenzyme to activate transcription [2]. In this manner, regulons or genes involved in the uptake and utilization of carbon source, flagellum synthesis, iron uptake, glycogen synthesis, outer membrane proteins etc. are regulated in E. coli and Salmonella Typhimurium [1, 22]. Moreover, insertional mutations in crp and cya have been reported to affect the production of proteins exported by the Ysc (Yersinia secretion), Ysa (Yersinia secretion apparatus), and flagellar T3SS in Yersinia enterocolitica [29]. Nevertheless, little was known regarding the role of CRP in Salmonella virulence until a recent study reported that crp/cya are required for expression of sirA gene which is an upstream regulator for Salmonella pathogenicity island 1 (SPI-1) gene clusters [37].
The SPI-1 T3SS is one of the virulence determinants of Salmonella that is important for intestinal colonization and invasion of epithelial cells as a prelude to systemic infection. By delivering more than 10 different effector proteins into the host cells via SPI-1 T3SS, Salmonella actively invades intestinal mucosa and causes enteropathy in the host [31, 42]. Moreover, the SPI-1 T3SS effector protein SipB induces caspase-1 dependent apoptosis in macrophages and plays an essential role in Salmonella pathogenesis [16, 25, 26]. Therefore, the aim of this study was to investigate the functional relationship between CRP and SPI-1 T3SS in Salmonella Choleraesuis using a crp mutant (∆crp) and to better understand the basis of attenuation in the mutant strain.
2. MATERIALS AND METHODS
2.1. Bacterial strains, plasmids and media
A wild type (WT) strain of Salmonella enterica serovar Choleraesuis (No. 13312) and an E. coli K-12 strain (No. 10798) were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). Bacteria were cultured at 37 °C in Luria-Bertani (LB) broth. To induce the expression of SPI-1 T3SS genes in the experiments described, bacteria were cultured at 37 °C in LB supplemented with 0.3 M NaCl under conditions of low aeration [14, 28, 36]. Where appropriate, ampicillin (100 μg/mL), kanamycin (50 μg/mL) or chloramphenicol (12.5 μg/mL) were added to the medium. Quantification of viable bacteria, expressed as colony forming units (CFU) per mL of culture or g of tissue, was determined for each experiment by preparation of serial dilutions of cultures or tissue homogenates in phosphate buffered saline (PBS) and plating on MacConkey or brilliant green (BG) agar plates (Becton, Dickinson and Company, Franklin Lakes, NJ, USA).
2.2. Construction of crp gene mutants
Mutants were generated by allelic exchange between suicide vectors and bacterial genomic DNA using a method described previously [40]. The mutant strains generated in this study are listed in Table I. Briefly, genomic DNA of Salmonella Choleraesuis was PCR amplified using the following primer pair: 5′- cat taa aat ctg gta ccg gag gag gac tct c -3′ and 5′- gat gaa ttc cca cca gca gaa gtt gcg tta cca -3′. The resulting 1 318 bp PCR product containing the full length crp (633 bp) flanked by partial sequences of the putative inner membrane protein genes, yhfA and yhfK, was digested with KpnI and EcoRI, ligated into pGP704, and maintained in the permissive E. coli strain SY327λpir [24]. The resulting plasmid was termed pGCRP. Thereafter, a HincII DNA fragment harboring a kanamycin-resistance determinant (km) was subsequently cloned and inserted into the crp gene of pGCRP. The resulting plasmid, pGCRPK, was a suicide vector for allelic exchange of crp by transformation into Salmonella Choleraesuis via electroporation (1.25 kV/cm, 600 Ω and 25 μF). The resulting bacterium CP327 strain exhibiting kanamycin-resistance and ampicillin-sensitivity was isolated and characterized by Southern hybridization using DNA probes specific to crp and km.
Table I.
Bacterial strains used in this study.
| Strain | Genotype | Relevant characteristics | Source |
|---|---|---|---|
| Salmonella enterica serovar Choleraesuis | |||
| WT | Wild type | ATCC, No. 13312 | |
| CP327 | crp-327::km | Derived from WT, KmR | This study |
| CP359 | crp-359::loxP | Derived from WT | This study |
| PQ518 | phoQ518::km | Derived from WT, KmR | This study |
| Escherichia coli | |||
| K12 | Wild type | ATCC, No. 10798 | |
In order to construct a crp mutant free of the km resistance gene, similar cloning procedures were applied. Briefly, a km determinant flanked by two direct repeats of the loxP sequence (5′- ata act tcg tat aat gta tgc tat acg aag tta t -3′) was cloned into the crp gene of pGCRP. The resulting suicide vector was transformed and resulting mutants with kanamycin-resistance and ampicillin-sensitivity were selected and genotypically characterized. The selected strain was a crp mutant harboring a loxP-flanked-kanamycin determinant. To excise the km gene, bacteria were further transformed with a temperature sensitive p705-Cre plasmid that harbored a Cre recombinase gene (Gene Bridges GmbH, Dresden, Germany). Excision of the km determinant from genomic DNA of candidate strains via Cre-loxP recombination was achieved by shifting the incubation temperature from 30 °C to 42 °C [44]. The increased temperature induced expression of Cre recombinase and simultaneously inhibited replication of p705-Cre, leading to the elimination of the plasmid. The resulting bacterium CP359 strain lacking an antibiotic gene was further identified by Southern hybridization analysis (data not shown).
2.3. Construction of plasmids encoding SopB, SipB and CRP
To construct a plasmid that encoded the N-terminal Myc-tagged SopB, genomic DNA of Salmonella Choleraesuis was amplified by PCR using primer pair sopB P1(F) and sopB P2(R) as shown in Table II. The resulting sopB gene was restriction digested with BglII and HindIII, and ligated into a pUC19 vector (GenBank Accession L09137) that harbored a promoter region for the neomycin determinant of the Tn5 transposon (GenBank Accession U00004). The resulting pTSopB plasmid contains the sopB-myc gene that is transcriptionally regulated by the neomycin promoter in cis. Similar procedures were used in the construction of plasmids pTSipB and pTCRP expressing N-terminal Myc-tagged SipB and CRP, respectively, using the pUC19 vector as a backbone (Tab. II).
Table II.
Oligonucleotide primers used and plasmids constructed in this study.
| Primer | Sequences (5′ → 3′)a | Plasmid |
|---|---|---|
| sopB P1(F) | CAGAGATCTGATCAAGAGACAGGATGAGGATCGTTTCGCATGCAAATACAGAGCTTC | pTSopB |
| sopB P2(R) | CCGAAGCTTTTACAGATCCTCTTCAGAGATGAGTTTCTGCTCAGATGTGATTAATGAAGA | |
| sipB P1(F) | CAGAGATCTGATCAAGAGACAGGATGAGGATCGTTTCGCATGGTAAATGACGCAAGTAGCATTAG | pTSipB |
| sipB P2(R) | CCGCTGCAGTTACAGATCCTCTTCAGAGATGAGTTTCTGCTCTGCGCGACTCTGGCGCAGAATAAAACG | |
| crp P1(F) | CAGAGATCTGATCAAGAGACAGGATGAGGATCGTTTCGCATGGTGCTTGGCAAACC | pTCRP |
| crp P2(R) | CCGAAGCTTTTACAGATCCTCTTCAGAGATGAGTTTCTGCTCACGGGTGCCGTAGACGA | |
| crp P3(F) | AATGCCGGATCCTGGACAGCAAGCGAACCGGAATTGCC | p705CRP |
| crp P3(R) | CGGGGATCCTCACACAGGAAACAGCTATGAC |
The Myc-tag sequences are shown in italics. Restriction sites are underlined. Stop codons are shown in bold italics. Sequences of partial neomycin promoter of Tn5 transposon are shown in bold.
Additionally, a plasmid that harbored the pSC101 replication of origin and the crp gene was constructed. Briefly, a p705 M plasmid that served as the backbone was generated from modification of p705-Cre by BamHI digestion and self-ligation. To clone crp into p705 M, the crp gene was PCR amplified using primers crp P3(F) and crp P3(R) with pTCRP as the template. The PCR product was restriction digested with BamHI and ligated into p705 M. The plasmid was termed p705CRP.
2.4. Complementation experiments
Complementation experiments were performed by transformation of pTCRP or p705CRP plasmids into CP327 strain via electroporation (1.25 kV/cm, 600 Ω and 25 μF). In the experiments of in vitro survival and LDH assay, the pTCRP plasmid was used. The secretory function of the complemented Salmonella Choleraesuis ∆crp was investigated by co-transformation of pTSopB and p705CRP into CP327. These two plasmids are compatible in the CP327 strain.
2.5. In vitro survival assay
Stationary phase cultures of Salmonella in LB were washed twice with PBS, and resuspended in control media. The control medium was M9 (pH 7.0) for the acid killing assay and saline (0.9% NaCl, pH 7.0) for the bile salts killing assay. Approximately 1 × 108 CFU of bacteria were inoculated into the M9 medium (pH 3.0), 15% bile salts in saline (Becton, Dickinson and Company), 1% sodium deoxycholate in saline (Becton, Dickinson and Company), or control medium, followed by gentle mixing and incubation at 37 °C for 1 h. After incubation, viable bacterial counts were determined on MacConkey agar plates. The percentage of surviving bacteria was calculated as the number of viable bacteria before incubation/the number of viable bacteria after incubation.
2.6. Secretion assay and Western blot analysis
Salmonella strains harboring appropriate plasmid(s) were cultured into LB broth containing 0.3 M NaCl and grown at 37 °C overnight. The bacterial pellets were collected by centrifugation at 3 000 × g for 10 min, resuspended in the same medium to ~ 1 × 109 CFU/mL and incubated at 37 °C for an additional 3 h. Thereafter, bacterial pellets were collected by centrifugation and culture supernatants were passed through a filter with a pore-size of 0.45 μm (Millipore, Billerica, MA, USA). All samples were normalized to the OD595 nm values determined at harvest and subjected to Western blot analysis. The protein bands on immunoblots recognized by anti-Myc antibody (Abcam, Cambridge, UK) represented the quantity of recombinant proteins present within bacterial cells or that were secreted by bacteria into culture supernatants. Signals from immunoblots probed with an anti-DnaK antibody (Merck, Darmstadt, Germany) represented the quantity of bacteria in pelleted fractions or in the supernatants containing lysed cells. The intensity of the bands was quantified by analysis of the scanned immunoblots using ImageJ software, version 1.37v (National Institutes of Health).
2.7. Animal experiments
The animals used in this study were cared for and handled according to the guidelines approved by the Institutional Animal Care and Use Committee (IACUC) of the National Chung Hsing University (permit number 94-67).
2.7.1. LD50 in mice
Healthy female BALB/c mice at 6 to 8 weeks of age were purchased from the National Laboratory Animal Center, Taiwan. The virulence of the Salmonella Choleraesuis strains was determined using both peroral (p.o.) and intraperitoneal (i.p.) routes of administration. For p.o. inoculations, mice were deprived of food and water for 4 h and orally given 30 μL of 10% sodium bicarbonate (Sigma, St. Louis, MO, USA) 5 min before administration of 40 μL of serially diluted Salmonella Choleraesuis in PBS. Food and water were resupplied 30 min after p.o. inoculation of bacteria. Intraperitoneal inoculation of mice was performed by injecting 100 μL of a bacterial suspension into the abdomen [9]. Mice that appeared severely ill were euthanized. Mortality was recorded at 21 days post inoculation and the LD50 was calculated by the method of Reed and Muench [30].
2.7.2. Porcine ligated ileal loop
Male Landrace × Yorkshire (LY) pigs at 8 weeks of age were obtained from a local commercial supplier. Pigs were confirmed to be negative for Salmonella prior to experiments by overnight enrichment of rectal swabs in Rappaport broth (Becton, Dickinson and Company) at 37 °C, followed by culture on BG agar plates overnight. For surgical manipulations, pigs were sedated by intramuscular administration of 4 mg/kg Stresnil (Janssen Animal Health, High Wycombe, UK), followed by induction of anesthesia by intravenous administration of 8 mg/kg thiamylal sodium (Shinlin Sinseng Pharmaceutical, Taiwan, R.O.C.), and maintenance of anesthesia with 2% isoflurane (Halocarbon Laboratories, River Edge, NJ, USA) in oxygen via an endotracheal tube. When a deep stage of anesthesia was reached, a midline incision was made through the abdominal wall, the ileocecal junction was revealed, and sequential 6-cm loops with 1-cm spacers were constructed with surgical silk. Twelve ileal loops were constructed in each pig and randomly assigned for bacterial inoculation. Bacteria grown under conditions to induce SPI-1 T3SS were injected into assigned loops at a concentration of 1 × 109 CFU/5 mL per loop. At least triplicate loops were injected for each bacterial strain. The abdomen was then closed with surgical thread and anesthesia was maintained for 3 h to allow intestinal invasion of bacteria. Thereafter, animals were euthanized and the ileal loops accompanied with the associated mesenteric lymph nodes were harvested.
To quantify the invasion of Salmonella, the ileal loops were dissected and gently washed with 250 mL of ice-cold saline to remove mucus and excess bacteria in the lumen. The intestinal walls with Peyer patches were harvested and were homogenized in PBS containing 0.5% bovine serum albumin (Sigma) and 0.5% Tergitol TM10 (Fluka, Buchs, Switzerland). Serial dilutions of homogenates of the intestine or mesenteric lymph nodes were plated on BG agar with appropriate antibiotic and incubated overnight at 37 °C. Bacterial counts in each preparation were performed as described previously.
2.8. Invasion and intracellular survival assays
Porcine alveolar macrophages (PAM) were obtained from 3- to 6-week old healthy piglets through lavage and stored in liquid nitrogen as previously described [5, 18]. For experiments, PAM were thawed, resuspended and cultured in RPMI-1640 (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin, and 2.5 μg/mL amphotericin B. Invasion and survival assays were performed as described previously with minor modifications [10, 12]. Briefly, PAM were plated in 96-well tissue culture plates at a density of 2 × 105 cells per well and incubated at 37 °C in 5% CO2 overnight. Fifty microliters of SPI-1 T3SS induced bacteria in culture medium (RPMI-1640 supplemented with 10% FBS) were added to PAM in each well to provide a final multiplicity of infection (MOI) of ~ 20. The plates were then centrifuged at 167 × g for 5 min and incubated at 37 °C in 5% CO2 to allow invasion. Two hours post inoculation (hpi), the cells were washed five times with culture medium and incubated in 200 μL of culture medium supplemented with 100 μg/mL gentamicin (Gibco Labs, Grand Island, NY, USA) for an additional 2 h in order to kill extracellular bacteria. Subsequently, the medium was replaced with culture medium containing 10 μg/mL gentamicin. At 4, 8 and 16 hpi, the cells were washed once with culture medium and lysed in 200 μL of 0.1% Triton X-100 in PBS for 5 min. Cell lysates were diluted serially in PBS and plated onto BG agar plates to determine the number of bacteria surviving intracellularly. The invasiveness of each strain was calculated as follows: CFU at 4 hpi/total bacterial counts at 2 hpi, and the relative invasion rate was expressed after normalization to WT. The survival rate of each strain at 8 or 16 hpi was calculated as (CFU at 8 or 16 hpi/CFU at 4 hpi) × 100%.
2.9. LDH assay
Murine RAW 264.7 macrophage cells were maintained in RPMI-1640 supplemented with 10% FBS. Bacteria grown under conditions that induce SPI-1 T3SS were used [28]. To determine the cytotoxic effect of Salmonella, RAW 264.7 macrophage cells were seeded onto 96-well cell culture plates at a density of 5 × 104 cells/well and incubated at 37 °C overnight. The cells were washed with RPMI-1640 and incubated for 1 h in RPMI-1640 containing 50 μM of the caspase-1 inhibitor Ac-YVAD-CMK (Calbiochem, Darmstadt, Germany) or the vehicle (0.5% dimethylsulfoxide, DMSO) used to dissolve the caspase-1 inhibitor. Bacterial suspensions representing an MOI of ~ 50 were added to the cell cultures and incubated for 1 h at 37 °C. Thereafter, low serum media with gentamicin (RPMI-1640 containing 1% FBS, 100 μg/mL gentamicin, and 50 μM Ac-YVAD-CMK or 0.5% DMSO) was added to kill extracellular bacteria and the cultures were incubated for an additional 6 h. Cytotoxicity was quantified using a kit (Roche, Basel, Switzerland) that detects LDH released from the cytoplasm. Percent cytotoxicity was calculated as 100 × ((experimental release − spontaneous release)/(total release − spontaneous release)), in which the spontaneous release was the amount of LDH activity in the supernatant of uninfected cells and the total release of LDH was measured from cells lysed with 1% Triton X-100 (Sigma).
3. RESULTS
3.1. The Salmonella Choleraesuis ∆crp was attenuated in BALB/c mice
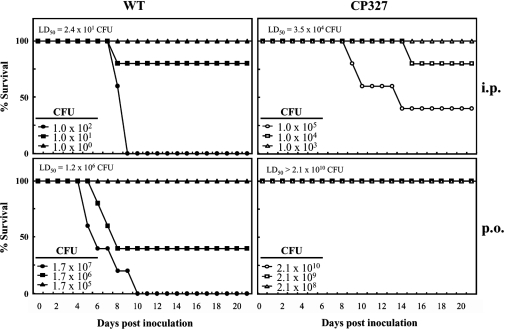
To examine the role of the crp gene in the virulence of Salmonella Choleraesuis, BALB/c mice were infected by p.o. or i.p. administration of the WT strain of Salmonella Choleraesuis or an isogenic crp gene mutant strain CP327. The results showed that the WT strain was highly virulent with an LD50 of 2.4 × 101 CFU and 1.2 × 106 CFU via delivery by i.p. and p.o. routes, respectively. Mice infected with the WT isolate developed acute systemic symptoms and death was observed within 10 days post inoculation (Fig. 1). By contrast, CP327 was greatly attenuated with LD50 doses of 3.5 × 104 CFU (i.p.) and > 2.1 × 1010 CFU (p.o.), that were approximately 103- and 104-fold greater than those for the WT, respectively (Fig. 1). Mice inoculated with CP327 showed a high tolerance for infection by the mutant strain and survived longer compared with those infected with WT, i.e. 40% of mice survived at 2 weeks after i.p. injection with 1 × 105 CFU of ∆crp. Notably, all mice challenged with 2.1 × 1010 CFU of CP327 delivered p.o. survived throughout the experimental period, indicating that CP327 was highly attenuated when administered via this route.
Figure 1.
Salmonella Choleraesuis ∆crp is attenuated in BALB/c mice. Survival of mice was recorded up to 3 weeks after peroral (p.o.) or intraperitoneal (i.p.) administration of varying doses of Salmonella Choleraesuis (WT) or the CP327 strain. Data are representative of two independent experiments (n = 5 in each group).
3.2. Attenuation of ∆crp was not due to greater susceptibility to low pH or bile salts
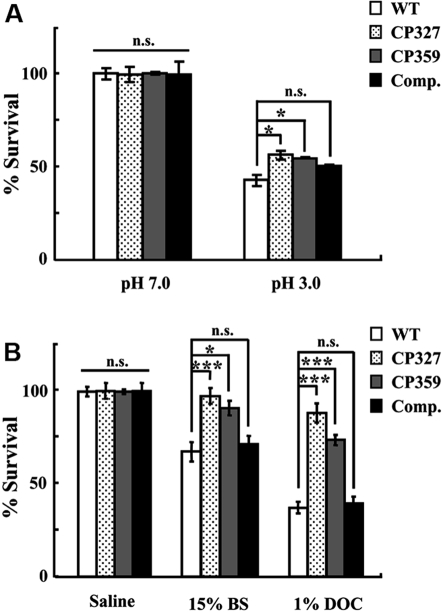
The fecal-oral route is the primary pathway for transmission of Salmonella infections among swine [11]. Before intestinal colonization, Salmonella needs to overcome the bactericidal effects of gastric acid and bile salts present in the upper alimentary tract. To determine if the attenuation of the ∆crp mutant was due to increased susceptibility to these bactericidal factors, survival of bacteria under conditions mimicking gastric acid and bile were investigated. The results show that both WT and ∆crp (CP327 and CP359) were highly sensitive to low pH (pH 3.0) with nearly 50% of bacteria being killed after 1 h incubation (Fig. 2A). However, the ∆crp strains showed ~ 10% higher resistance than WT (p < 0.05) indicating that ∆crp strains were less susceptible to acidic environments. Furthermore, incubation of WT in the presence of 15% bile salts or 1% sodium deoxycholate (an active component of bile) resulted in the inactivation of nearly 30% and 60% of the bacteria, respectively (Fig. 2B). By contrast, ∆crp strains were significantly more resistant to both reagents than the WT and the sensitivity of the CP327 was restored by complementation in trans with a plasmid encoding CRP (Figs. 2A and 2B).
Figure 2.
Salmonella Choleraesuis ∆crp is more resistant to low pH or bile salts. Bacteria grown to stationary phase in LB broth were incubated for 1 h in M9 medium at pH 3.0 or 7.0 (A) or in saline, 15% bile salts, or 1% sodium deoxycholate (B). Survival of bacteria before and after incubation was determined on MacConkey agar plates. Percent survival was normalized to the initial inoculum. WT, wild type Salmonella Choleraesuis; CP327 and CP359, crp gene mutants; Comp., CP327 complemented with pTCRP plasmid; BS, bile salts; DOC, sodium deoxycholate. The results are from three independent experiments of at least triplicate determinations. Error bars are SEM. *p < 0.05; ***p < 0.001; n.s.: no significant difference from each other (ANOVA).
3.3. Intestinal invasion was impaired in ∆crp
During infection, Salmonella Choleraesuis must penetrate the intestinal epithelial barrier prior to establishing systemic infection. To examine the role of the crp gene in intestinal invasiveness, an in vivo experiment was performed using porcine ligated ileal loops. The result shows that, 3 h after incubation, fewer colonies of CP327 were recovered from the ileal wall and associated Peyer patches as compared to WT (p < 0.05). Further examination of the Salmonella strains that had penetrated the intestinal epithelial barrier revealed that significantly fewer CP327 were recovered from the mesenteric lymph nodes as compared to WT (p < 0.001) (Fig. 3A) suggesting that the mutation in the crp gene was associated with reduced intestinal colonization and/or invasion of Salmonella Choleraesuis.
Figure 3.
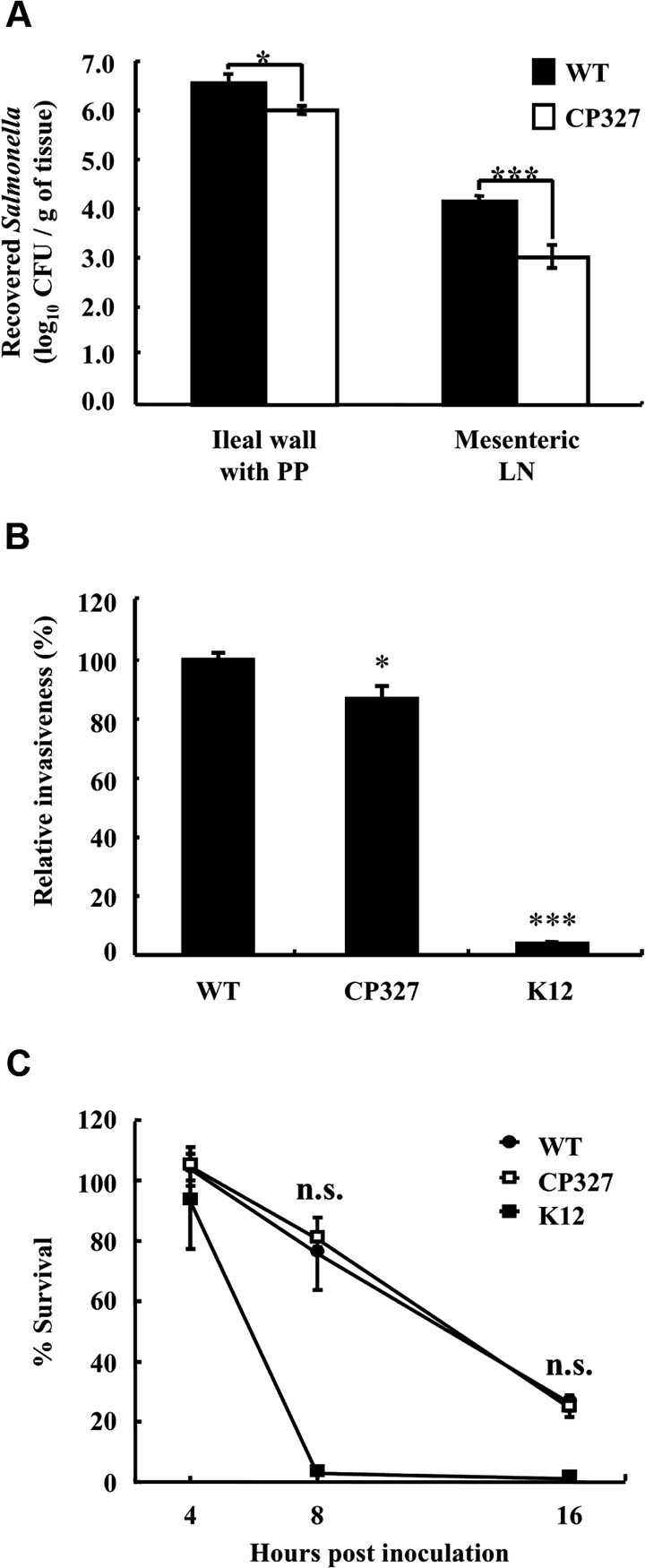
Salmonella Choleraesuis ∆crp was less invasive in porcine ligated ileal loops and in macrophages. The degree of intestinal invasion by wild type Salmonella Choleraesuis (WT) and CP327 was determined at 3 h after inoculation of the loops (A). To recover bacteria, portions of the ileal wall with Peyer patches (PP) and associated mesenteric lymph nodes (LN) were harvested, homogenized, and plated onto BG agar. The results are from at least triplicate determinations. For in vitro invasion (B) and intracellular survival assays (C), porcine alveolar macrophages were co-cultured with WT and CP327 of Salmonella or E. coli (K12) strains at an MOI of ~ 20 for 2 h and intracellular survived bacteria were assessed. Relative invasion was determined at 4 hpi while intracellular survival rates were at 8 and 16 hpi. The results are representatives of three independent experiments of at least triplicate determinations. Error bars are SEM. *p < 0.05 and ***p < 0.001 compared to WT; n.s.: no significant difference from each other (ANOVA).
Salmonella is able to invade and survive within macrophages that in turn serve as vehicles for bacteria to spread systemically. To determine the basis for our observation that fewer numbers of CP327 were present in mesenteric lymph nodes in ligated ileal loop experiments, we performed in vitro assays of bacterial invasion and survival in PAM. The results show that, compared to the WT, mutations in crp resulted in a 10% reduction in invasiveness of CP327 (Fig. 3B). These results can be compared with those from co-cultivation of PAM with a non-invasive strain of E. coli (K12) where only 4% of the viable bacteria were recovered, representing a background level of bacterial phagocytosis by PAM. In comparing the bacterial survival rate within PAM, it was noted that both Salmonella strains, but not E. coli K12, were able to survive within PAM. However, there was no significant difference between WT and CP327. At 16 hpi, the percent of viable Salmonella strains decreased to approximately 25% (Fig. 3C).
3.4. Secretion of SPI-1 effector proteins SopB and SipB was impaired in ∆crp
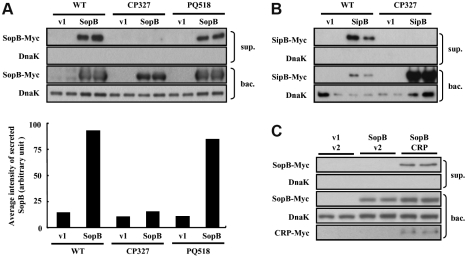
The SPI-1 encoded T3SS delivers a set of effector proteins which facilitates bacterial entry into intestinal epithelial cells [45]. To understand whether a mutation in the crp gene affected SPI-1 T3SS and contributed to the impaired intestinal invasiveness of Salmonella Choleraesuis, the secretory function of SPI-1 T3SS was investigated by ectopic expression of the Myc-tagged recombinant effector proteins SopB and SipB in WT and mutant strains. It was observed that, under conditions that induced SPI-1 T3SS, WT bacteria successfully expressed the recombinant protein SopB and secreted it into the supernatant (Fig. 4A). To investigate whether the presence of SopB in culture supernatants was attributed to spontaneous bacterial lysis, the leakage of DnaK, a cytoplasmic chaperon protein, was examined by probing the same immunoblot with a DnaK-specific antibody. The lack of a DnaK signal in the bacterial culture supernatant confirmed that SopB was indeed secreted and not released due to bacterial lysis. By contrast, recombinant SopB was ectopically expressed in CP327, however, secretion of this protein was not detectable in the culture supernatant (Fig. 4A). To exclude the possibility that the altered function of SPI-1 T3SS was due to genetic manipulations used during the construction of CP327, an isogenic phoQ gene mutant PQ518 strain, constructed using a similar strategy, was tested for SopB secretion. The results showed that the secretory phenotype of PQ518 was comparable to that of the WT strain (Fig. 4A) suggesting that the genomic manipulation procedures used in this study did not affect SPI-1 T3SS.
Figure 4.
Secretion of effector proteins SopB and SipB via SPI-1 T3SS was impaired in ∆crp. Wild type (WT), CP327 or PQ518 strains transformed with pUC19 (v1), pUC19 encoding sopB-myc (SopB), sipB-myc (SipB), p705 M (v2) or p705 M encoding crp-myc (CRP) were grown under conditions that induced SPI-1 T3SS function. Bacterial culture supernatants (sup.) and pellets (bac.) were harvested, normalized and subjected to Western blot analysis with an antibody to Myc-tag. The results of the SopB secretion assay in WT, CP327 and PQ518 strains (A), SipB secretion assay in WT and CP327 (B) and SopB secretion assay in CP327 complemented with p705CRP (C). Immunoblots were also probed with an anti-DnaK antibody as a loading control and to detect signals resulting from bacterial lysis. The average intensity of secreted proteins was quantified and normalized to the intensity of DnaK for WT, CP327 and PQ518 strains (A, lower panel).
To further demonstrate that the impairment of SPI-1 T3SS was a general effect and not restricted to SopB, secretion of another effector protein SipB was also assessed (Fig. 4B). Recombinant SipB was secreted efficiently in WT bacteria. However, there was no detectable signal in the supernatant of CP327 cultures even though a large amount of SipB was expressed within the CP327 cells (Fig. 4B). Furthermore, complementation of CP327 with co-expressed recombinant CRP restored the ability of the mutant to secrete SopB extracellularly, while complementation with a control plasmid remained non-secretory (Fig. 4C).
3.5. Caspase-1-dependent cytotoxicity in RAW 264.7 macrophage cells was reduced in ∆crp
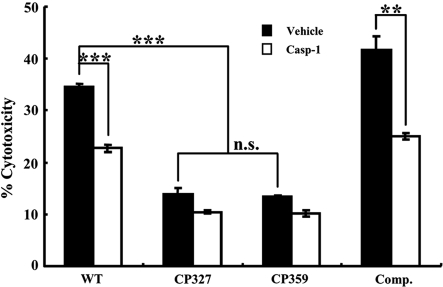
Salmonella is known to activate caspase-1-dependent cytotoxicity in macrophages by delivering the SPI-1 T3SS effector protein SipB to the cell membrane [25, 38]. To determine whether there was impaired secretion of SipB by ∆crp affected macrophage cytotoxicity, RAW 264.7 cells were co-cultured with Salmonella at an MOI of ~ 50 for 6 h in the presence of the caspase-1-specific inhibitor Ac-YVAD-CMK or DMSO and subjected to cytotoxicity analysis using an LDH release assay. The results showed that WT Salmonella Choleraesuis induced approximately 35% cytotoxicity in RAW 264.7 cells and the addition of Ac-YVAD-CMK significantly reduced cytotoxicity to 23% (Fig. 5). By contrast, cytotoxicity caused by CP327 or CP359 strains was only ~ 14%, indicating that ∆crp mutants were less cytotoxic to macrophages (Fig. 5). To further confirm that the reduction in cytotoxicity was attributed to the mutation of the crp gene, the effect of CRP complementation was investigated. It was noted that when complemented with a plasmid expressing CRP, the CP327 strain regained the ability to induce cytotoxicity in macrophages and the cytotoxicity was reduced significantly by a caspase-1 inhibitor Ac-YVAD-CMK (p < 0.01) (Fig. 5).
Figure 5.
Salmonella Choleraesuis induced caspase-1 dependent cytotoxicity in RAW 264.7 cells was impaired in ∆crp mutants. Bacteria grown under conditions that induce SPI-1 T3SS were co-cultured at a MOI of ~ 50 with RAW 264.7 cells treated with 0.5% DMSO carrier (Vehicle) or with 50 μM caspase-1 inhibitor Ac-YVAD-CMK (Casp-1). Cell death was assessed at 6 hpi by measuring the release of lactate dehydrogenase (LDH). WT, wild type Salmonella Choleraesuis; CP327 and CP359, crp gene mutants; Comp., CP327 complemented with pTCRP plasmid. The results are from three independent experiments of at least triplicate determinations. Error bars are SEM. **p < 0.01; ***p < 0.001; n.s.: no significant difference from each other (ANOVA).
4. DISCUSSION
The application of live Salmonella vaccines as a preventive measure has been effective in the control of swine salmonellosis [32]. By eliciting effective humoral and cellular immune responses in the vaccinated animals, live Salmonella Choleraesuis vaccines harboring a mutation in crp have been shown to induce protective immunity in mice and pigs [8, 20]. One of the characteristics of such mutants is the attenuation in virulence due to unknown mechanism(s). In this study, we showed that, at the early stage of infection, the attenuation in ∆crp was not attributed to increased susceptibility to low pH or bile salts, but to a reduction in invasiveness associated with an impairment of the secretory function of SPI-1 T3SS and lower invasion rate and cytotoxicity to macrophages. This study is the first to demonstrate that a mutation in the crp gene leads to a functional defect in the SPI-1 T3SS and to delineate the role of CRP in Salmonella Choleraesuis virulence.
Genes targeted for the development of live Salmonella vaccines fall into two major categories, metabolic and virulence-associated genes, of which, the former were found to be more immunogenic [23]. Nevertheless, not all mutants deficient in a metabolic-gene are attenuated or effective in inducing immunity. For example, in contrast to the findings in Salmonella Typhimurium [17], Nnalue et al. (1987) reported that Salmonella Choleraesuis strains with mutations in the aroA (aromatic biosynthesis) or galE (UDP-glucose-4-epimerase) genes were attenuated, but did not induce a protective response in BALB/c mice [27]. Distinctively, mutations in the crp gene of Salmonella enterica serovars Typhimurium, Choleraesuis, or Typhi lead to the attenuation of yielding strains that may serve as effective vaccine candidates in mice, pigs, horses or human beings [8, 20, 33, 35, 43]. However, it is not clear how crp mutants display reduced virulence yet manage to elicit a protective host immune response.
Using ligated ileal loops of pigs, we found that intestinal invasion of Salmonella Choleraesuis ∆crp was significantly impaired as compared to WT. Nevertheless, there were still approximately 106 CFU (per gram of tissue) of ∆crp attached to or invaded into the ileum wall, and 103 CFU (per gram of tissue) of ∆crp were able to reach mesenteric lymph nodes, suggesting that the ability of ∆crp to invade intestinal tissues was not fully abolished and that a pathway independent of SPI-1 T3SS might have allowed Salmonella-infected phagocytes to gain access and spread to mesenteric lymph nodes, where the immune response occurs. This hypothesis is supported by other investigations showing that Salmonella Typhimurium deficient in SPI-1 also gained access to the spleen from the intestine [13] and that CD18-expressing phagocytes are responsible for the extra-intestinal dissemination of Salmonella [39]. Therefore, it is plausible that Salmonella Choleraesuis crp mutants might also invade and subsequently induce immune responses in the spleen and/or lymph nodes via a similar pathway. Moreover, because SPI-1 T3SS activity was impaired in Salmonella Choleraesuis ∆crp, we speculate that the immunogenicity of Salmonella Choleraesuis does not rely on a functional SPI-1 T3SS. This hypothesis is supported by a recent study that demonstrated that protection against Salmonella Typhimurium in pigs is independent of SPI-1 T3SS secreted proteins [3].
Survival within macrophages is an essential feature of Salmonella pathogenesis in vivo [12]. In this study, we found that the ∆crp mutant was less invasive for PAM as compared to WT, but the survival rates were similar. Therefore, a reduction in the invasiveness of the ∆crp mutant probably accounts for the reduced number of ∆crp cells observed in mesenteric lymph nodes of ligated ileal loops. Other investigators have demonstrated decreased colonization of Peyer patches and spleens of mice infected by Salmonella Choleraesuis strains having mutations in the crp/cya genes. Moreover, adhesion/invasion in CHO cells and bacterial transcytosis in polarized Madin–Darby canine kidney cells was significantly reduced as compared to WT [19]. Taken together, these findings strengthen our results that suggest these phenotypes are correlated to impaired SPI-1 T3SS function in Salmonella Choleraesuis ∆crp.
However, CRP is pleiotropic and mutations in the crp gene have been shown to alter diverse characteristics of Salmonella including carbon source utilization, synthesis of flagella, reduced growth rate, and attenuation in virulence [9, 43]. Due to the versatile role of CRP, we cannot exclude the possibility that virulence determinants other than SPI-1 T3SS may also be affected in Salmonella Choleraesuis ∆crp. For example, a recent report indicated that there is a significant decrease of SPI-2 gene expression in the crp mutant of Salmonella Typhimurium [41]. Also, our results show that a mutation in the crp gene increased the resistance of Salmonella Choleraesuis to low pH or bile salts, and ∆crp was more resistant to gentamicin as comparing to the parental strain (data not shown). These data led us to hypothesize that a mutation in crp might have effects on the composition or structure of the cell wall resulting in decreased permeability to bile salts or gentamicin.
In conclusion, this study explored the gastrointestinal factors that affect Salmonella Choleraesuis ∆crp colonization/invasion and provides evidence to delineate one of the attenuation mechanisms of Salmonella Choleraesuis ∆crp that is widely approached as a candidate of live Salmonella vaccine.
Acknowledgments
This work was supported in part by the Bureau of Animal and Plant Health Inspection and Quarantine, Council of Agriculture, Taiwan, Republic of China and by the Division of Animal Health, Bayer HealthCare.
References
- 1.Botsford J.L., Harman J.G., Cyclic AMP in prokaryotes, Microbiol. Rev. (1992) 56:100–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Busby S., Ebright R.H., Transcription activation by catabolite activator protein (CAP), J. Mol. Biol. (1999) 293:199–213 [DOI] [PubMed] [Google Scholar]
- 3.Carnell S.C., Bowen A., Morgan E., Maskell D.J., Wallis T.S., Stevens M.P., Role in virulence and protective efficacy in pigs of Salmonella enterica serovar Typhimurium secreted components identified by signature-tagged mutagenesis, Microbiology (2007) 153:1940–1952 [DOI] [PubMed] [Google Scholar]
- 4.Chang C.C., Lin Y.H., Chang C.F., Yeh K.S., Chiu C.H., Chu C., et al. , Epidemiologic relationship between fluoroquinolone-resistant Salmonella enterica serovar Choleraesuis strains isolated from humans and pigs in Taiwan (1997 to 2002), J. Clin. Microbiol. (2005) 43:2798–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chien M.S., Chan Y.Y., Chen Z.W., Wu C.M., Liao J.W., Chen T.H., et al. , Actinobacillus pleuropneumoniae serotype 10 derived ApxI induces apoptosis in porcine alveolar macrophages, Vet. Microbiol. (2009) 135:327–333 [DOI] [PubMed] [Google Scholar]
- 6.Chiu C.H., Wu T.L., Su L.H., Chu C., Chia J.H., Kuo A.J., et al. , The emergence in Taiwan of fluoroquinolone resistance in Salmonella enterica serotype Choleraesuis, N. Engl. J. Med. (2002) 346:413–419 [DOI] [PubMed] [Google Scholar]
- 7.Chiu C.H., Chuang C.H., Chiu S., Su L.H., Lin T.Y., Salmonella enterica serotype Choleraesuis infections in pediatric patients, Pediatrics (2006) 117:e1193–e1196 [DOI] [PubMed] [Google Scholar]
- 8.Chu C.Y., Wang S.Y., Chen Z.W., Chien M.S., Huang J.P., Chen J.J., et al. , Heterologous protection in pigs induced by a plasmid-cured and crp gene-deleted Salmonella choleraesuis live vaccine, Vaccine (2007) 25:7031–7040 [DOI] [PubMed] [Google Scholar]
- 9.Curtiss R. III, Goldschmidt R.M., Fletchall N.B., Kelly S.M., Avirulent Salmonella typhimurium ∆cya ∆crp oral vaccine strains expressing a streptococcal colonization and virulence antigen, Vaccine (1988) 6:155–160 [DOI] [PubMed] [Google Scholar]
- 10.Elsinghorst E.A., Measurement of invasion by gentamicin resistance, Methods Enzymol. (1994) 236:405–420 [DOI] [PubMed] [Google Scholar]
- 11.Fedorka-Cray P.J., Kelley L.C., Stabel T.J., Gray J.T., Laufer J.A., Alternate routes of invasion may affect pathogenesis of Salmonella typhimurium in swine, Infect. Immun. (1995) 63:2658–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fields P.I., Swanson R.V., Haidaris C.G., Heffron F., Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent, Proc. Natl. Acad. Sci. USA (1986) 83:5189–5193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galan J.E., Curtiss R. III, Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells, Proc. Natl. Acad. Sci. USA (1989) 86:6383–6387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galan J.E., Curtiss R. III, Expression of Salmonella typhimurium genes required for invasion is regulated by changes in DNA supercoiling, Infect. Immun. (1990) 58:1879–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hackett J., Salmonella-based vaccines, vaccine (1990) 8:5–11 [DOI] [PubMed] [Google Scholar]
- 16.Hersh D., Monack D.M., Smith M.R., Ghori N., Falkow S., Zychlinsky A., The Salmonella invasin SipB induces macrophage apoptosis by binding to caspase-1, Proc. Natl. Acad. Sci. USA (1999) 96:2396–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoiseth S.K., Stocker B.A., Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines, Nature (1981) 291:238–239 [DOI] [PubMed] [Google Scholar]
- 18.Iglesias G., Pijoan C., Molitor T., Interactions of Pseudorabies virus with swine alveolar macrophages I: virus replication, Arch. Virol. (1989) 104:107–115 [DOI] [PubMed] [Google Scholar]
- 19.Kelly S.M., Bosecker B.A., Curtiss R. III, Characterization and protective properties of attenuated mutants of Salmonella choleraesuis , Infect. Immun. (1992) 60:4881–4890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kennedy M.J., Yancey R.J. Jr, Sanchez M.S., Rzepkowski R.A., Kelly S.M., Curtiss R. III, Attenuation and immunogenicity of ∆cya ∆crp derivatives of Salmonella choleraesuis in pigs, Infect. Immun. (1999) 67:4628–4636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ku Y.W., McDonough S.P., Palaniappan R.U., Chang C.F., Chang Y.F., Novel attenuated Salmonella enterica serovar Choleraesuis strains as live vaccine candidates generated by signature-tagged mutagenesis, Infect. Immun. (2005) 73:8194–8203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawson C.L., Swigon D., Murakami K.S., Darst S.A., Berman H.M., Ebright R.H., Catabolite activator protein: DNA binding and transcription activation, Curr. Opin. Struct. Biol. (2004) 14:10–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meeusen E.N., Walker J., Peters A., Pastoret P.P., Jungersen G., Current status of veterinary vaccines, Clin. Microbiol. Rev. (2007) 20:489–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller V.L., Mekalanos J.J., A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR , J. Bacteriol. (1988) 170:2575–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monack D.M., Raupach B., Hromockyj A.E., Falkow S., Salmonella typhimurium invasion induces apoptosis in infected macrophages, Proc. Natl. Acad. Sci. USA (1996) 93:9833–9838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monack D.M., Hersh D., Ghori N., Bouley D., Zychlinsky A., Falkow S., Salmonella exploits caspase-1 to colonize Peyer’s patches in a murine typhoid model, J. Exp. Med. (2000) 192:249–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nnalue N.A., Stocker B.A., Test of the virulence and live-vaccine efficacy of auxotrophic and galE derivatives of Salmonella choleraesuis, Infect. Immun. (1987) 55:955–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel J.C., Galan J.E., Investigating the function of Rho family GTPases during Salmonella/host cell interactions, Methods Enzymol. (2008) 439:145–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petersen S., Young G.M., Essential role for cyclic AMP and its receptor protein in Yersinia enterocolitica virulence, Infect. Immun. (2002) 70:3665–3672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reed L.J., Muench H., A simple method of estimating fifty percent endpoints, Am. J. Hyg. (1938) 27:493–497 [Google Scholar]
- 31.Schlumberger M.C., Hardt W.D., Salmonella type III secretion effectors: pulling the host cell’s strings, Curr. Opin. Microbiol. (2006) 9:46–54 [DOI] [PubMed] [Google Scholar]
- 32.Schwartz K.J., Salmonellosis, in: Straw B.E., D’Allaire S., Mengeling W., Taylor D.J. (Eds.), Disease of swine, 8th ed., Iowa State University Press, Ames, 1999, pp. 535–551 [Google Scholar]
- 33.Sheoran A.S., Timoney J.F., Tinge S.A., Sundaram P., Curtiss R. III, Intranasal immunogenicity of a ∆cya ∆crp-pabA mutant of Salmonella enterica serotype Typhimurium for the horse, Vaccine (2001) 19:3787–3795 [DOI] [PubMed] [Google Scholar]
- 34.Stabel T.J., Mayfield J.E., Morfitt D.C., Wannemuehler M.J., Oral immunization of mice and swine with an attenuated Salmonella choleraesuis [∆cya-12 ∆(crp-cdt)19] mutant containing a recombinant plasmid, Infect. Immun. (1993) 61:610–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tacket C.O., Hone D.M., Curtiss R. III, Kelly S.M., Losonsky G., Guers L., et al. , Comparison of the safety and immunogenicity of ∆aroC ∆aroD and ∆cya ∆crp Salmonella typhi strains in adult volunteers, Infect. Immun. (1992) 60:536–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tartera C., Metcalf E.S., Osmolarity and growth phase overlap in regulation of Salmonella typhi adherence to and invasion of human intestinal cells, Infect. Immun. (1993) 61:3084–3089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teplitski M., Goodier R.I., Ahmer B.M., Catabolite repression of the SirA regulatory cascade in Salmonella enterica, Int. J. Med. Microbiol. (2006) 296:449–466 [DOI] [PubMed] [Google Scholar]
- 38.Van der Velden A.W., Lindgren S.W., Worley M.J., Heffron F., Salmonella pathogenicity island 1-independent induction of apoptosis in infected macrophages by Salmonella enterica serotype Typhimurium, Infect. Immun. (2000) 68:5702–5709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vazquez-Torres A., Jones-Carson J., Baumler A.J., Falkow S., Valdivia R., Brown W., et al. , Extraintestinal dissemination of Salmonella by CD18-expressing phagocytes, Nature (1999) 401:804–808 [DOI] [PubMed] [Google Scholar]
- 40.Yeh K.S., Hancox L.S., Clegg S., Construction and characterization of a fimZ mutant of Salmonella typhimurium, J. Bacteriol. (1995) 177:6861–6865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoon H., McDermott J.E., Porwollik S., McClelland M., Heffron F., Coordinated regulation of virulence during systemic infection of Salmonella enterica serovar Typhimurium, PLoS Pathog. (2009) 5:e1000306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang S., Kingsley R.A., Santos R.L., Andrews-Polymenis H., Raffatellu M., Figueiredo J., et al. , Molecular pathogenesis of Salmonella enterica serotype Typhimurium-induced diarrhea, Infect. Immun. (2003) 71:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X., Kelly S.M., Bollen W.S., Curtiss R. III, Characterization and immunogenicity of Salmonella typhimurium SL1344 and UK-1 ∆crp and ∆cdt deletion mutants, Infect. Immun. (1997) 65:5381–5387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y., Buchholz F., Muyrers J.P., Stewart A.F., A new logic for DNA engineering using recombination in Escherichia coli, Nat. Genet. (1998) 20:123–128 [DOI] [PubMed] [Google Scholar]
- 45.Zhou D., Galan J., Salmonella entry into host cells: the work in concert of type III secreted effector proteins, Microbes Infect. (2001) 3:1293–1298 [DOI] [PubMed] [Google Scholar]