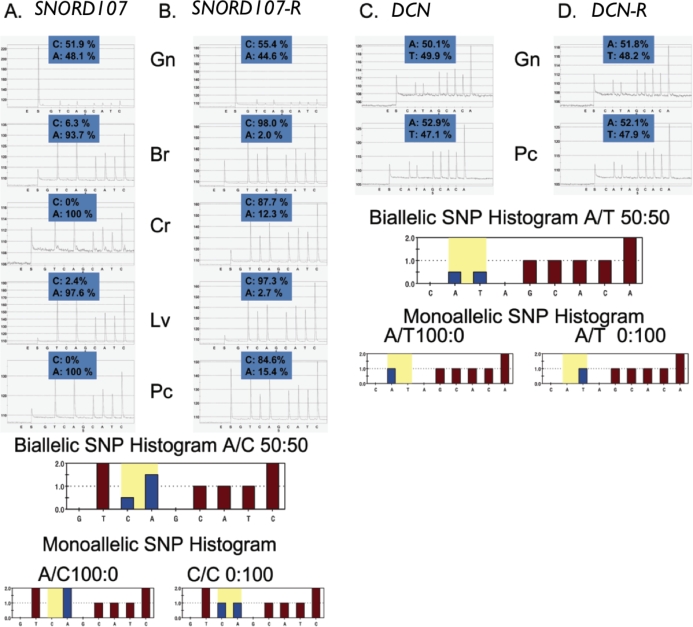
FIG. 9.
QUASEP results of Day 30 fetal samples obtained from reciprocal Meishan × WC matings. A representative set of pyrosequencing results for both (A and B) imprinted (SNORD107) and (C and D) nonimprinted (DCN) genes. Reciprocal matings (R) from WC × Meisan and Meishan × WC for SNORD107 (B) and DCN (D) are included. Allelic quantification of each pyrogram can be calculated on the basis of peak height and the data used to test for bias from the genomic sample. DCN was not expressed on microarrays in brain or liver tissues, and carcass assays were not available. The biallelic and monoallelic histograms represent the ideal expected peak heights that would correspond for each particular tSNP. Neighboring sequence may affect interpretation of expected peak height ratios, as depicted in the SNORD107 pyrograms (i.e., TTcA looks different than TTCA). Additionally, to account for any PCR bias, the genomic and cDNA results are shown for all tissues and each assay. The numbers above the peaks denote the percentage of each allele identified by QUASEP. In all cases, they should add up to 100%. Multiple replications of this QUASEP assay were used to develop the results shown in Table 2. Gn, genomic DNA sample; Br, brain cDNA; Cr, carcass cDNA; Lv, liver cDNA; Pc, placenta cDNA.