Table 1.
Rate and equilibrium constants for TRAPP and nucleotide binding to Ypt1p.
| Parameter | Value | Source |
|---|---|---|
| GDP | ||
| k+1 | 0.096 (±0.004) μM-1 s-1 | mant-dGDP binding |
| 0.05 (±0.01) μM-1 s-1 | mantGDP binding10 | |
| k-1 | 0.00012 (±0.00002) s-1 | [H3]GDP Filter binding10 |
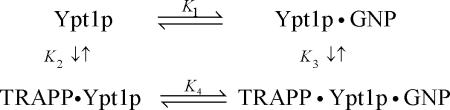
| K1 | 1.2 (±0.2) nM | Ratio of rate constants |
| K3 | 4.8 (±1.2) μM | mant-GDP/mant-dGDP (Figure 1) |
| k+4A | 2.4 (±0.1) μM-1 s-1 | Linear fit (Figure 2) |
| 2.5 (±0.1) μM-1 s-1 | Global fit (Figure 2) | |
| k-4A | 22.1 (±2.0) s-1 | Linear fit (Figure 2) |
| 15.7 (±5.3) s-1 | Global fit (Figure 2) | |
| k+4B | 20.7 (±2.2) s-1 | Hyperbolic fit (Figure 2) |
| 11.7 (±12.0) s-1 | Global fit (Figure 2) | |
| k-4B | 0.17 (±0.01) s-1 | mantGDP/mant-dGDP (Figure 1) |
| K4A = (k-4A/k+4A) | 9.2 (±0.9) μM | Ratio of rate constants |
| 6.3 (±2.1) μM | Ratio of rate constants (global fit) | |
| K4B = (k-4B/k+4B) | 0.01 (±0.001) | Ratio of rate constants |
| K4,overall | 75 (±10) nM | Calculated using Eq. 5 |
| K2 | 77 (±25) nM | Detailed balance of Scheme 1 |
| GTP/GMPPNP | ||
| k+1 | 0.10 (±0.01) μM-1 s-1 | mantGTP binding10 |
| k-1 | 0.00005 s-1 | GTP and mant-GMPPNP dissociation11 |
| K1 | 0.5 (±0.1) nM | Ratio of rate constants |
| K3 | 8 (±2) μM | mant-GMPPNP (Figure 1) |
| k-4 | 0.095 (±0.017) s-1 | mant-GMPPNP (Figure 1) |
| k+4 | 3.2 (±0.1) μM-1 s-1 | mantGTP binding10 |
| K4 | 30 (±5) nM | Ratio of rate constants |
| K2 | 133 (±48) nM | Detailed balance of Scheme 1 |
