Abstract
RNA interference (RNAi) can be an effective antiviral agent; however, overexpression of RNAi can be toxic through competition with the endogenous microRNA (miRNA) machinery. We used rational design to identify highly potent RNAi that is effective at nontoxic doses. A statistical analysis was conducted to pinpoint thermodynamic characteristics correlated with activity. Sequences were selected that conformed to a consensus internal stability profile (ISP) associated with active RNAi, and RNAi triggers were expressed in the context of an endogenous miRNA. These approaches yielded highly active hepatitis B virus (HBV) RNAi. A statistical analysis found a correlation between activity and nucleation by binding within the seed sequence to accessible regions in the target RNA. Guide strands were selected for favorable strand biasing, but increased strand biasing did not correlate with potency, suggesting a threshold effect. Exogenous short hairpin RNAs (shRNAs), but not miRNAs were previously reported to compete with miRNAs for the miRNA/RNAi machinery. In contrast, we show that exogenous Polymerase III- but not Polymerase II-driven miRNAs compete with exogenous miRNAs, at multiple steps in the miRNA pathway. Exogenous miRNAs also compete with endogenous miR-21. Thus, competition with endogenous miRNAs should be monitored even when using miRNA-based therapeutics. However, potent silencing was achieved at doses where competition was not observed.
Introduction
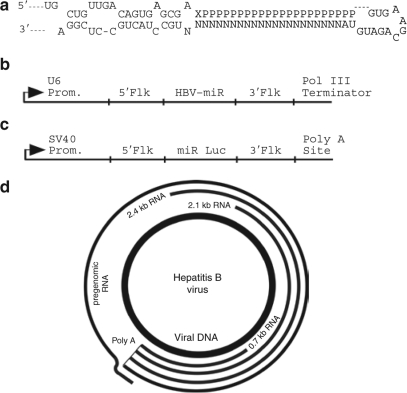
Hepatitis B virus (HBV) is a DNA virus that replicates through an RNA intermediate. HBV infection is strongly associated with hepatocellular carcinoma1 and is the ninth leading cause of death worldwide according to the World Health Organization.2 Current treatments are costly, have significant side effects3,4 and are effective in only ~50% of patients.5 HBV produces four major classes of mRNAs. The approximate sizes of these mRNAs are 3.5, 2.4, 2.1, and 0.7 kb. The 3.5 kb pregenomic RNA is the template for replication of the viral DNA minus strand. The HBV genome contains extensive overlapping reading frames (Figure 1d).
Figure 1.
Genomic organization of hepatitis B virus (HBV) and structure of microRNA (miRNA) scaffold. (a) miR-30 scaffold for RNAi expression. N indicates nucleotides within miR-30 that were substituted with HBV guide strands. P indicates residues complementary to the guide strand that constitute the passenger strand. X indicates a nucleotide complementary to the 21st nucleotide of the guide strand. (b) Structure of the Pol III expression cassette used to express HBV-miRNAs and miR Luc. 5′Flk and 3′Flk indicate endogenous miR-30 5′ and 3′ flanking sequences, respectively. Prom, promoter. (c) Structure of SV40 miR Luc. (d) HBV has four overlapping reading frames. All of the RNAi triggers target the largest (pregenomic) RNA. A subset of the RNAi triggers target the 2.4 + 2.1 kb RNAs.
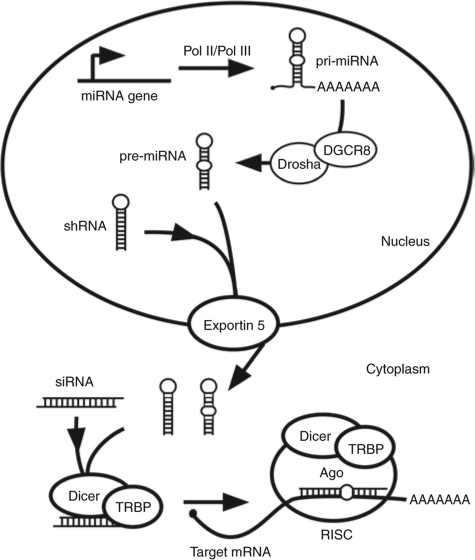
RNA interference (RNAi) is an endogenous gene-silencing pathway that responds to double-stranded (ds)RNAs by silencing homologous genes.6 Specific silencing of a transcript can be directed by the transfection of cells or animals with short-interfering RNAs (siRNAs, Figure 2). Alternatively, short hairpin RNAs (shRNAs) can be expressed from plasmid or viral templates within cells or animals to trigger silencing (Figure 2). Recently, it has been appreciated that the RNAi machinery is also involved in normal gene regulation. MicroRNAs (miRNAs) are a family of ~22 nucleotide single-stranded noncoding RNAs that silence endogenous transcripts (reviewed in ref. 7). miRNAs are synthesized as part of a hairpin structure embedded within a primary miRNA (pri-miRNA, Figure 2). In the nucleus, Drosha and DGCR8 (DiGeorge syndrome critical region gene 8) cooperate to cleave pri-miRNAs into hairpin-like structures called pre-miRNAs. It is likely that exogenous shRNAs are substrates for the RNAi/miRNA machinery because they resemble pre-miRNAs. Pre-miRNAs are transported to the cytoplasm by Exportin-5 (ref. 8) and cleaved into mature miRNAs by Dicer. Mature miRNAs form duplexes with endogenous target RNAs and either prevent their translation or cause their degradation.7 Again, it is likely that synthetic siRNAs are substrates for the RNAi/miRNA machinery because they resemble the duplex produced by Dicer cleavage of pre-miRNAs.
Figure 2.
The miRNA/RNAi pathway. Pri-miRNAs are processed by Drosha/DGCR8 into pre-miRNAs. Pre-miRNAs and shRNAs are transported to the cytoplasm by Exportin-5 where they are processed into siRNAs by Dicer. A single strand of the siRNA is incorporated into RISC, which silences genes by translational inhibition or target mRNA cleavage. shRNAs and siRNAs enter the pathway at indicated steps. DGCR8, DiGeorge syndrome critical region gene 8; miRNA, microRNA, pri-miRNA, primary miRNA; RISC, RNA-induced silencing complex; RNAi, RNA interference; shRNA, short hairpin RNA; siRNA, short-interfering RNA.
RNAi has been used to inhibit a wide variety of viruses in cultured cells and in vivo. Previously, we developed a transient model for testing HBV RNAi-based therapeutics in which HBV replication and RNAi were initiated by co-transfection of cultured hepatoma cells or mouse liver with HBV and RNAi expression plasmids encoding shRNAs.9 Using this model, we and subsequently others, have shown that RNAi could be used to inhibit HBV replication (reviewed in ref. 10). Because no rational design rules were available at the time of our initial study, RNAi sequences were chosen based on conservation of sequences among the major HBV genotypes. The most potent construct identified in this screen was HBVU6#2, which reduced the levels of HBV RNAs by 77% in mice.
In a subsequent study, Grimm et al. expressed HBVU6#2 from an adeno-associated virus (AAV) serotype 8 (ref. 11). Treatment of HBV transgenic mice with high doses of this virus encoding HBVU6#2 resulted in substantial reductions in HBV RNAs. However, this vector, as well as numerous other vectors encoding other shRNAs, was lethal within one month. Lethality coincided with decreases in the levels of abundant miRNAs in the liver (miR-122 and let-7a). The RNAi and miRNA pathways share many components. The authors suggested that the toxicity was due to competition of the shRNA for the endogenous miRNA machinery. Because overexpression of Exportin-5 (the factor responsible for nuclear export of miRNA precursors) increased silencing, the authors reasoned that Exportin-5 might be saturable in their system. This sobering report highlights the need to develop the most potent RNAi-inducing molecules (RNAi triggers) such that low doses, which do not saturate the endogenous miRNA machinery, can be used. It is clearly also important to monitor saturation of the endogenous miRNA machinery when testing novel therapeutic RNAi.
HBVU6#2 was identified in a nonrational screen. In several other screens, 1/3–1/4 of tested RNAi triggers were functional.12,13,14 Since the design of our original constructs, several reports have emerged that suggest approaches for rational design of more potent RNAi.15,16,17,18 Using our HBV model system, we show here that a combination of these rational approaches can be utilized to identify highly potent RNAi triggers with high frequency. We also conducted a statistical analysis to identify factors that correlate with potency. Lastly, therapeutic RNAi must be safe as well as potent. Because HBVU6#2 was previously shown to compete for the endogenous miRNA machinery, we examined the ability of exogenous HBV-miRNAs to compete with exogenous siRNAs, shRNAs, and miRNAs as well as an endogenous miRNA, miR-21.
Results
Design of more potent RNAi triggers
To determine whether rational design would lead to more potent RNAi, 14 RNAi sequences were selected (Table 1) using a combination of four design guidelines described in the following sections, which were available at the start of this study. Predicted Dicer cleavage products were based on miR-30 mapping studies by Lee et al.19
Table 1.
RNAi sequences, targets and locations in HBV genome
Expression of RNAi sequences in the context of an endogenous miRNA. RNAi triggers were embedded within the context of the endogenous miRNA, miR-30, with endogenous flanking sequences, and a U6 promoter (Figure 1a,b). It was reported that expression in a miRNA context led to a more fully processed guide strand and more potent silencing than expression in an shRNA context.15
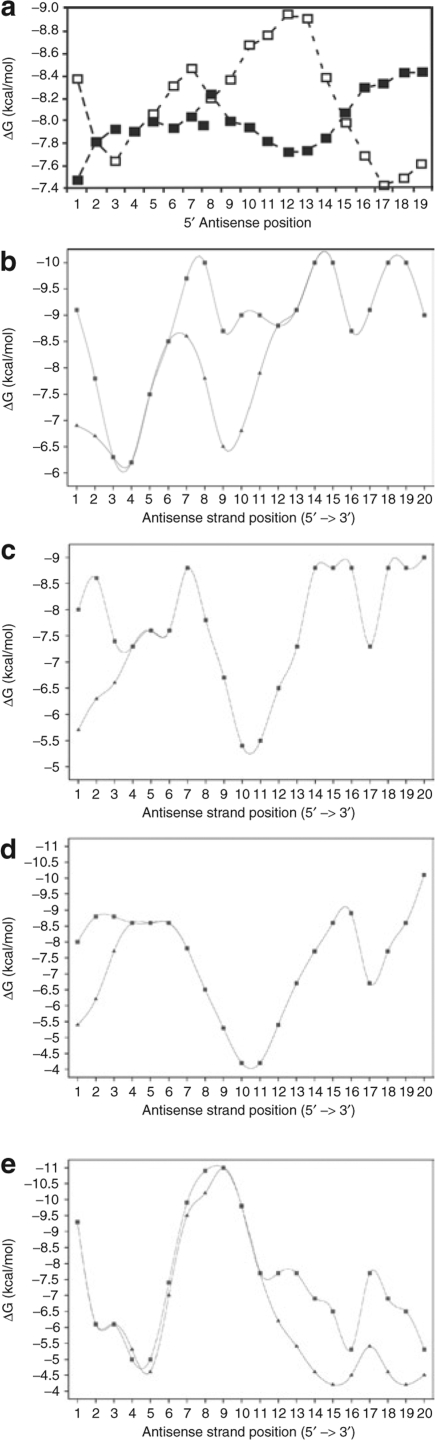
Strand asymmetry. Asymmetric stability of the siRNA influences which strand enters the RNA-induced silencing complex (RISC).16,17 Functional siRNAs and endogenous miRNAs tend to have: (i) weak pairing of the 5′ antisense strand base pairs, (ii) weak pairing of bases 11–14, and (iii) strong pairing of the 3′ antisense base pairs.16 The algorithm, Sfold,20 was used to identify sequences that most closely conformed to the consensus thermodynamic internal stability profile (ISP) identified by Khvorova et al.16 (Figure 3a). Figure 3b–d (squares) shows the calculated ISP for four triggers used in this article and Figure 3e shows the ISP for HBVU6#2. The ISPs of the remaining constructs used in this article are shown in Supplementary Figures S1–S12. None of the sequences examined exactly matched the consensus ISP, so GU wobbles were introduced by mutating the passenger strand to improve the ISP (Table 1, Figure 3b–e, and Supplementary Figures S1–S13, diamonds).
Figure 3.
The thermodynamic profile of an siRNA determines RISC strand bias. (a) Calculated average internal stability profiles for a set of 180 siRNAs targeting every other position of firefly luciferase and human cyclophilin. (Adapted with permission from ref. 16). Predicted internal stability profiles of three selected RNAi triggers: (b) HBV-miR 2, (c) HBV-miR 6, and (d) HBV-miR 10. Original profiles indicated by (filled squares). GU wobbles were introduced by mutation of the sense strand in order to force profiles to more closely conform to the consensus stability profile shown in a [indicated by (filled diamonds)]. Remaining internal stability profiles are shown in Supplementary Figures S1–S13. (e) Predicted internal stability profile for HBVU6#2. GU wobbles were introduced to facilitate cloning in bacteria, but were not intended to optimize the match with the consensus internal stability profile in the case of HBVU6#2. HBV, hepatitis B virus; RISC, RNA-induced silencing complex; RNAi, RNA interference; siRNA, short-interfering RNA.
Empirical sequence features. Based on a report by Reynolds et al., when possible, sequences containing a U in the sense strand at position 10 (HBV-miRs 2, 6, 10, 11, 12, and 14) were chosen and a G base at position 13 (all HBV-miRs except 4, 8, 13, and 14) was avoided.
Target site accessibility. At the time the RNAi triggers were designed, the role of target RNA secondary structure was controversial. Sfold was, therefore, used to assign an accessibility score to 19 mers within the HBV RNAs and four triggers that had good strand asymmetry and also had a high Sfold accessibility score based on the entire 19 mer target site were selected (HBV-miRs 5, 10, 11, and 12). In more recent analyses,21 structure-based energetic calculations were considered for more sophisticated modeling of the hybridization process (see section on Statistical analysis of parameters influencing extent of silencing).
The four major mRNAs of HBV share overlapping reading frames (Figure 1d). The largest RNA, termed the pregenomic RNA, is targeted by all the RNAi triggers. A subset of the RNAi triggers target the 2.4 and 2.1 kb pre-S1 and pre-S2/S transcripts. Because these transcripts overlap, quantification by quantitative PCR is problematic, so transcript levels were determined by northern blot throughout this article. Table 1 summarizes the RNAs targeted by the various RNAi triggers.
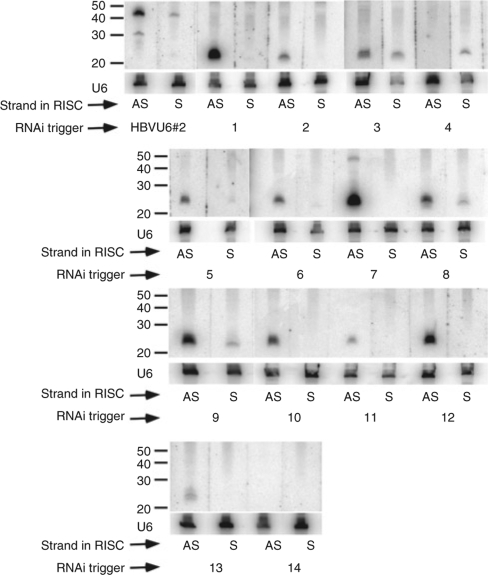
We first sought to determine whether selecting RNAi triggers that matched the consensus ISP resulted in preferential incorporation of the desired antisense strand into the RISC complex. Human hepatoma (Huh-7) cells were transfected with indicated HBV-miRNA or shRNA expression vectors and a plasmid containing 1.3 unit-length HBV genomes (pTHBV2). Transfection of hepatoma cells with pTHBV2 results in production of all four viral RNAs, synthesis of the four viral proteins, and production of replicated partially double-stranded HBV genomic DNA. After 3 days, RNAs were harvested and strand-specific northern blots were conducted to determine whether the desired antisense strand or undesired sense strand was incorporated into the RISC complex. Note that the passenger strand is cleaved upon RISC loading and is rapidly degraded. The U6 RNA was probed as a loading control. Figure 4 shows that for HBVU6#2, very little mature antisense guide strand is produced, while a significant amount of unprocessed precursor is observed. This is surprising because HBVU6#2 was the most potent RNAi trigger identified in our previous screen.9 Slightly more undesired mature sense vs. desired antisense guide strand was detected, indicating that HBVU6#2 is not optimally strand biased. For reasons that are not understood, neither the sense nor antisense strand was detected in cells transfected with HBV-miR 14. With the exception of HBV-miRs 3 and 4, preferential incorporation of the desired antisense strand into RISC was observed (Figure 4). In fact, for many of these HBV-miRNAs, there is no detectable sense strand. For HBV-miR-3, a mixture of sense and antisense was observed and HBV-miR-4 preferentially incorporated the undesired sense strand into RISC. Taken together, these results show that rational design leads to a high frequency of loading of the antisense strand into RISC (11 out of 13 times for which a sense or antisense guide strand was detected).
Figure 4.
Strand-specific small transcript northern blots show that rational design leads to incorporation of desired antisense into RISC in most cases. Small transcript northern blots were probed with strand-specific oligonucleotide probes. AS and S indicate that the antisense strand or sense strand, respectively, was incorporated into RISC. Blots were stripped and reprobed for the U6 RNA as a loading control. Molecular weight standards are indicated. HBV, hepatitis B virus; RISC, RNA-induced silencing complex; RNAi, RNA interference.
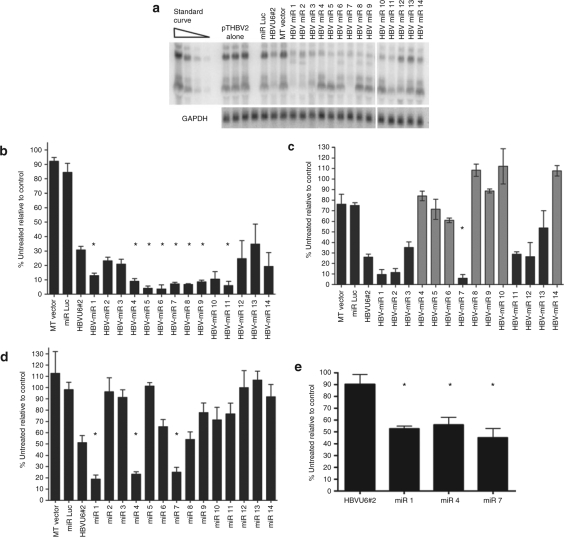
Silencing of HBV mRNAs. Next, the ability of the HBV-miRNA RNAi triggers to silence HBV RNAs was tested. As a basis for comparison, HBVU6#2, the most potent shRNA identified in our previous screen,9 was used. Huh-7 cells were transfected with 2 µg of the HBV genomic plasmid pTHBV2 (ref. 22), to initiate a viral replication cycle, as well as 2 µg of HBVU6#2 or HBV-miR expression plasmid. A northern blot was conducted and glyceraldehyde 3-phosphate dehydrogenase was used to normalize for loading. A representative northern blot is shown in Figure 5a. The average of three independent experiments is shown in Figure 5b,c. The percent inhibition was calculated relative to the average of three wells transfected with pTHBV2 alone. As discussed above, while all the HBV-miRs target the 3.5 kb HBV RNA, several do not target the 2.4 and 2.1 kb RNAs. Therefore, silencing of the 3.5 kb and the 2.4 + 2.1 kb RNAs was assessed in Figures 5b,c. All rationally designed HBV-miRNAs (HBV-miRs 1–14) triggered substantial silencing of HBV. At the 2 µg dose, HBVU6#2 reduced the levels of the 3.5 kb HBV RNA by 69.1%. In contrast, all the rationally designed RNAi triggers (with the exception of HBV-miR-13) showed a greater extent of silencing of the 3.5 kb RNA than HBVU6#2, with eight reaching statistical significance (Figure 5b, P < 0.05 indicated by an asterisk). As expected, HBV-miRNAs that do not target the 2.4 + 2.1 kb RNAs did not substantially reduce their levels (Figure 5c, gray bars, P > 0.33–1), while HBV-miRNAs that do target them reduced their levels (Figure 5c, black bars).
Figure 5.
Silencing of HBV RNAs. (a) A representative northern blot demonstrating silencing by HBV-miRNAs is shown. (b) Silencing of the 3.5 kb pgRNA at the 2 µg dose shows that eight miRNA-based RNAi triggers silence to a greater extent than the benchmark shRNA, HBVU6#2. All values are relative to cells transfected with pTHBV2 and stuffer plasmid. (c) Silencing of the 2.4 + 2.1 kb HBV RNAs at the 2 µg dose. Gray bars indicate HBV-miRNAs that do not target these RNAs, and as expected, do not significantly silence them. (d) Silencing of the 3.5 kb pgRNA at the 0.5 µg dose shows that three miRNA-based RNAi triggers silence to a greater extent than the benchmark shRNA, HBVU6#2. (e) Silencing of the 3.5 kb pgRNA at the 0.01 µg dose shows that three miRNA-based RNAi triggers retain potency, while HBVU6#2 does not. Asterisks indicate statistical significance (P < 0.05). Error bars indicate mean ± SD. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HBV, hepatitis B virus; miRNA, microRNA; RNAi, RNA interference; pgRNA, pregenomic RNA; shRNA, short hairpin RNA.
Because such a large extent of silencing was observed with some of the HBV miRNAs, it was difficult to determine the relative silencing efficiencies of these constructs. Therefore, silencing was examined as above but at a 0.5 µg dose (Figure 5d). HBVU6#2 reduced the levels of the 3.5 kb HBV RNA by 48.8%. HBV-miRs 1, 4, and 7 were significantly more potent, silencing the 3.5 kb RNA by 81.2, 76.7, and 74.8%, respectively (P < 0.05). As above, HBV-miRs that do not target the 2.4 + 2.1 kb RNAs (including HBV-miR 4) did not substantially reduce their levels (data not shown). Next, the ability of HBVU6#2, HBV-miR 1, HBV-miR 4, and HBV-miR 7 to silence at a 50-fold lower dose (0.01 µg) was tested. At this dose, the shRNA HBVU6#2 did not silence at all, while three HBV-miRs silenced by ~50% (Figure 5e, P < 0.05). Taken together, these data show that rational design can lead to the identification of highly potent RNAi triggers.
Statistical analysis of parameters influencing extent of silencing
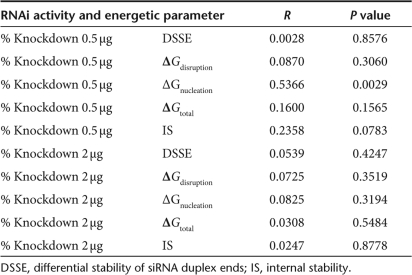
The moderate number of HBV RNAis tested in this article provided the opportunity to closely examine which thermodynamic characteristics of both the target mRNA and the siRNA duplexes were correlated with increased activity. A weighted regression analysis was conducted to identify correlations between various thermodynamic parameters discussed in the following paragraphs (Supplementary Table S1) and silencing efficiency at the 2 µg and 0.5 µg doses (Table 2). Khvorova et al. found that there was a correlation between siRNA activity and strand asymmetry.16 Biochemical studies by Schwarz et al.17 confirmed that strand asymmetry led to preferential incorporation of one strand into the RISC complex. The strand asymmetry was calculated as DSSE (differential stability of siRNA duplex ends), where DSSE measures the difference in stability between the two siRNA duplex ends (Supplementary Table S1, Materials and Methods). A guide strand has favorable strand asymmetry if DSSE > 0.0 kcal/mol. Consistent with Figure 4, the calculated DSSE for HBVU6#2 was unfavorable (DSSE < 0, Supplementary Table S1). Because all our RNAi triggers were designed for favorable strand asymmetry, all had DSSE > 0.0 kcal/mol. Interestingly, a weighted regression analysis showed that RNAi activity did not correlate with increasing DSSE, suggesting that once the rule of the siRNA duplex asymmetry is met, a larger difference in the stabilities between the siRNA ends does not lead to greater RNAi activity.
Table 2.
Weighted regression analyses for low dosage (0.5 µg) and high dosage (2 µg) for constructs with favorable DSSE
Recent studies on determinants for effective RNAi have suggested that target structural accessibility is important for silencing efficiency.21,23,24 mRNAs do not usually assume a single conformation, rather they exist as a population of structures. The Sfold algorithm generates a statistical sample of secondary structures from the Boltzmann ensemble of RNA secondary structures.20 We have previously undertaken a detailed thermodynamic approach to describe the interaction between guide strand and target in order to dissect out parameters that may be associated with siRNA efficacy21 and miRNA functionality.25 We proposed a model25 in which the first step in guide strand/target interaction is a nucleation step, during which a subset of bases binds to an unpaired stretch of the target mRNA. During this nucleation step, binding energy is gained through base pairing, but an energy cost is paid due to loss of translational and rotational entropy. ΔGnucleation represents the net energy gained by the nucleation event. For the interaction to be productive, ΔGnucleation must be <0. Unless the mRNA target site is fully unpaired, an energetic penalty must be paid to break the intramolecular target site base pairs (ΔGdisruption) so that the remaining base pairs of the guide strand/target can form. This is offset by new base pairs formed between the guide strand and the target RNA (ΔGhybrid). The net gain in energy upon binding is ΔGtotal = ΔGhybrid − ΔGdisruption. A weighted regression analysis was conducted to determine the correlation among ΔGnucleation, ΔGdisruption, or ΔGtotal and the extent of HBV silencing at the 2 and 0.5 µg doses. For simplicity, only silencing of the 3.5 kb RNA was considered.
Our statistical analysis found that nucleation energy was significantly correlated with the knockdown level at low dosage, with an R value of 0.5366 and a P value of 0.0029. Interestingly, for 7 of the 14 RNAi triggers, the predicted nucleation site resides within the target region that corresponds to the seed region (nucleotide 2–8 of the 5′ end) of the antisense siRNA. For 8 others, three of the four nucleating nucleotides reside within the seed-matching region. For HBVU6#2, the predicted nucleation site is completely outside the seed-matching region on the target. An appreciable but insignificant correlation for ΔGdisruption and ΔGtotal (Table 2) was observed. A likely reason for the lack of significance is proposed in the Discussion section. The consensus profile described by Khvorova et al. showed a decrease in stability at nucleotides 11–14. At the low dosage (0.5 µg), a relatively small correlation of 0.2358 was observed for this internal stability, with P value of 0.0783, just above the 0.05 cutoff (Table 2).
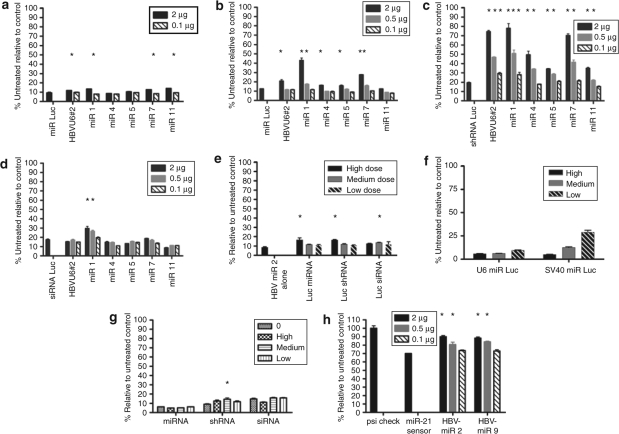
Competition between RNAi triggers: any therapeutic RNAi must not only be potent but also safe. At high doses, shRNAs can be toxic due to competition with the endogenous miRNA machinery. We sought to determine whether the use of miRNA expression vectors eliminates or reduces this type of toxicity. In a previous study, HBVU6#2, when expressed at high doses from an adeno-associated virus, appeared to inhibit the biogenesis of the endogenous liver miR-122 in mice.11 A recent report from the Rossi laboratory showed that both exogenous siRNAs and shRNAs could compete with the endogenous miRNA miR-21 (ref. 26). In contrast, a synthetic exogenous Pol II-driven miRNA did not compete with another synthetic exogenous miRNA or the endogenous miR-21. We concluded that the miRNA scaffold might be the safest expression context for RNAi. To determine whether HBVU6#2 or our HBV-miRs were saturating the miRNA machinery in cultured cells, the following series of experiments was conducted (Figure 6a–e). First, a synthetic Pol III-driven miR Luc, which targets firefly luciferase, was used as a surrogate for endogenous miRNAs. Cells were transfected with a luciferase expression plasmid, and either a high dose (1.5 µg, Figure 6a) or moderate dose (0.5 µg, Figure 6b) of miR Luc. Indicated doses of either HBVU6#2 shRNA or HBV-miRs 1, 4, 5, 7, or 11 were co-transfected. If the shRNA or HBV-miRs compete with miR Luc for the silencing machinery, the degree of luciferase silencing should decrease. At the higher dose of miR Luc, a small but statistically significant amount of competition by HBVU6#2 and several HBV-miRs (Figure 6a) was observed. The competition became more pronounced when the lower dose of miR Luc was used (Figure 6b). To our surprise, the amount of competition for two of the HBV-miRs (1 and 7) was greater than that seen with HBVU6#2 (P < 0.05). These data suggest that an exogenous miRNA mimic can compete with another exogenous miRNA mimic.
Figure 6.
Competition for miRNA/RNAi machinery by shRNAs or miRNA RNAi triggers. Cells were transfected with 0.5 µg of the luciferase expression plasmid pGL3-control (Promega) as well as indicated competitor (a–d and g). Competition with a (a) high dose (1.5 µg) or (b) low dose (0.5 µg) of a luc miRNA. Competition with a (c) luc shRNA or (d) luc siRNA. Legend indicates the dose of competitor. shRNAs and HBV-miRNAs compete with miRNAs and shRNAs. HBV-miRNAs compete with siRNAs. (e) Competition of luc miRNAs, shRNAs, and siRNAs against HBV miR-2. The amount of transfected RNAi trigger was adjusted to account for differences in molecular weight. High, moderate, and low doses used in micrograms were as follows: miRNA, 2, 0.5, and 0.1; shRNA, 2, 0.5, and 0.1; siRNA 0.21, 0.053, and 0.01. (f) Comparison of silencing efficiency of Pol III-driven U6 miR Luc and Pol II-driven SV40 miR Luc shows that SV40 miR Luc is less potent. The amount of transfected RNAi trigger was adjusted for molecular weight. High, medium, and low doses were the following (in micrograms): U6 miR Luc, 2, 0.5, and 0.1; SV40 miR Luc, 2.7, 0.7, and 0.13. (g) Competition of exogenous Pol II-driven SV40 HBV-miR 2 with an exogenous miRNA Luc, shRNA,Luc or siRNA Luc. Doses used were as in e. (h) HBV-miRNAs compete with endogenous miR-21. Cells were co-transfected with a miR-21 sensor that contains a bulged miR-21 binding site in the 3′ untranslated region of the renilla luciferase gene of psicheck 2 as well as indicated amounts HBV-miR 2 or HBV-miR 9. Asterisks indicate statistical significance (P < 0.05). Error bars indicate mean ± SD. HBV, hepatitis B virus; miRNA, microRNA; RNAi, RNA interference; shRNA, short hairpin RNA.
Next, a series of experiments were conducted to determine at which step in the silencing process the competing miR (or shRNA) was exerting its effect (see Figure 2). Competition could occur with: (i) Drosha/DGCR8 during conversion of the pri-miRNA to the pre-miRNA, (ii) with nuclear export by Exportin-5, (iii) Dicer/TRBP (TAR RNA-binding protein) cleavage of shRNAs or miRNAs to siRNAs or (iv) loading into RISC or Argonaute2 (Ago2) cleavage of target mRNAs. To eliminate some of these possibilities, an analogous set of experiments were conducted that measured competition against a luciferase shRNA or siRNA. If competition were occurring at step 1, one would not expect to see competition with a shRNA targeting luciferase. If competition were occurring at step 2 or 3, one would not expect to see competition with a siRNA targeting luciferase. In each case, the level of luciferase miRNA, shRNA, and siRNA was adjusted so as to give similar levels of luciferase silencing in the absence of competitor. As shown in Figure 6c, we observe robust competition with the luciferase shRNA by the HBVU6#2 shRNA as well as with our HBV-miRNAs. With some doses of shRNA and HBV-miR, silencing by the luciferase shRNA is almost completely abrogated. While this experiment does not exclude competition at step 1, it suggests that competition can occur downstream of Drosha/DGCR8. We next tested whether the HBV shRNA or HBV-miRs could compete with an exogenous siRNA. Figure 6d shows that at least HBV-miR 1 can also compete with a siRNA. This suggests competition at RISC loading or cleavage, although these experiments do not rule out competition at multiple steps. The fact that the same miR competes more with a luc shRNA than a luc miRNA supports competition at multiple steps, although an alternative interpretation is that shRNAs may in fact be poor substrates for the RNAi machinery. We then tested the ability of luc miRNAs, shRNAs, and siRNAs to compete against HBV-miR-2. Luc miRNAs, shRNAs, and siRNAs all showed significant competition with HBV-miR 2 (Figure 6e).
To determine whether competition would be observed when using a weaker Pol II promoter to drive expression of a miRNA-based trigger, we expressed miR Luc and HBV-miR 2 under the control of the SV40 promoter (Figure 1c). Figure 6f shows that SV40 miR Luc was less potent than the Pol III U6 miR Luc construct. We measured the ability of different doses of SV40 HBV-miR 2 to compete with a Luc miRNA, shRNA, or siRNA in experiments analogous to those shown in Figures 6b–d. With the exception of statistically significant competition with a shRNA at one dose of HBV-miR 2, this Pol II-driven miRNA did not compete at any step in the miRNA pathway. Thus, weaker Pol II-driven constructs appear to compete less with the endogenous miRNA machinery, consistent with the dose-dependent competition observed in Figures 6a–d.
We next sought to determine whether HBVU6#2 or our HBV-miRs might compete with an endogenous miRNA (miR-21) for the miRNA/RNAi machinery. To test for functional interference with miR-21, we constructed a reporter in which the binding site for miR-21 was cloned within the 3′ untranslated region of the renilla luciferase gene in the plasmid Psicheck 2 (Promega, Madison, WI). The firefly luciferase gene within Psicheck 2 serves as an internal control. This miR-21 binding site contains a bulge, such that cleavage of the renilla luciferase does not occur via RNAi. Rather, endogenous miR-21 binding silences renilla luciferase expression by ~30%, presumably by translational arrest or indirect transcript destabilization (Figure 6h). Cells were transfected with 2, 0.5, or 0.1 µg of HBV-miR 2 or HBV-miR 9. Statistically significant competition with endogenous miR-21 was observed with the 2 and 0.5 µg doses of HBV miRs 2 and 9 (Figure 6h). These data demonstrate that exogenous miRNA-based expression cassettes can compete with the function of an endogenous miRNA.
Discussion
Grimm et al. showed that high vector doses of expressed shRNAs could be fatal to mice.11 This toxicity was dose dependent. Thus, it would be desirable to design HBV RNAi triggers that are effective at lower doses. We, therefore, set out to rationally design HBV RNAi triggers expressed within the context of a natural miRNA.
All of our rationally designed RNAi triggers were designed to mimic the consensus thermodynamic profile described by Khvorova et al. Figure 3a).16 While none of the 3,182 potential sequences within the HBV genome exactly matched the consensus ISP, we were able to more closely match the ISPs by strategic introduction of GU wobbles. To our knowledge, this approach has not been reported previously. Strand-specific small transcript northern blots demonstrated that rational design led to preferred incorporation of the desired antisense strand into RISC in almost all cases.
Previously, we conducted a screen for effective shRNAs targeting HBV. Typically, ~25% of RNAi triggers tested in screens show substantial knockdown.12,13,14 Using rational design principles, we were able to select very potent RNAi triggers targeting HBV. All of our HBV-miRNA-based triggers showed substantial silencing and 8 out of 14 were more potent than our benchmark shRNA, HBVU6#2 (P < 0.05). At lower doses, three RNAi triggers were substantially more potent than HBVU6#2; this should allow inhibition of HBV using lower doses, thus minimizing toxicity.
We conducted a statistical analysis of the factors contributing to the potency of our RNAi triggers. Our data indicate that the nucleation step is important for efficient RNAi. The results on the predicted location of nucleation site support the hypothesis that the seed region is very likely to be involved in the initial binding by siRNA to an accessible nucleation site on the target. Interestingly, we found that increased strand biasing did not increase activity, suggesting a threshold effect. The results on internal stability indicate that, only at low dosage, low stability near the cleavage site may have some effect on RNAi efficiency.
We did not observe a statistically significant correlation between activity and ΔGdisruption or ΔGtotal. We note that the distribution of energy values is incomplete for either ΔGdisruption or ΔGtotal. Specifically, there is a lack of data for energy interval (−7.7 kcal/mol, 0.0 kcal/mol) for ΔGdisruption, or (−17.7 kcal/mol, 0.0 kcal/mol) for ΔGtotal (Supplementary Table S1). Thus, the dataset is not statistically representative for analyses on ΔGdisruption and ΔGtotal. This could bias the results and lead to the lack of significance in correlation for these parameters. Substantial differences in the correlations were observed between high siRNA dosage and low dosage. Incorporation of siRNA concentration presents a challenge for prediction of RNAi activity and the rational design of siRNAs and shRNAs.
Several reports have shown that siRNAs26,27,28,29 and shRNAs11,26 can compete with each other or with endogenous miRNAs for the miRNA/RNAi machinery. However, Castanotto et al. reported that Pol II-driven miRNA expression cassettes were safer than shRNAs because they did not compete with shRNAs or with endogenous miRNAs.26 Because HBVU6#2 had been shown to cause toxicity in mice due to competition with the endogenous RNAi machinery, we tested whether our HBV-miRNA triggers would also compete with other miRNAs. Our data (Figures 6a–d) show that miRNA-based RNAi triggers can compete moderately with another exogenous miRNA, highly with shRNAs and slightly with siRNAs. The competition we observe with the luc siRNA suggests that RISC loading or “slicer” activity is saturable. This is consistent with reports indicating that RISC activity is saturable in vitro30 as well as in cells.26 The fact that much greater competition against the shRNA was observed is consistent with saturation occurring at a second step such as the Exportin-5 or Dicer/TRBP step. Alternatively, the luc shRNA may be a poor substrate for the miRNA/RNAi machinery and is thus easy to compete with. From our data, we cannot exclude the possibility that the shRNA and the miRNAs competed at the Drosha/DGCR8 processing step. Castanotto et al. did not observe competition by an exogenous miRNA with exogenous shRNAs or Pol II-driven miRNAs.26 We observe competition when using miRNA expression cassettes driven by the powerful U6 Pol III promoter but not by the weaker SV40 promoter (Figures 6f,g). We also observe competition between exogenous HBV-miRs and endogenous miR-21 Figure 6h), which is a moderately abundant liver miRNA (data not shown).
Our data suggest that competition between exogenous miRNAs and endogenous miRNAs should be considered in future experiments when using miRNA-based vectors for RNAi expression. However, we still observe significant levels of silencing with our HBV-miR-based RNAi expression cassettes at doses where we do not observe significant competition with other miRNAs. Thus, miRNA-based RNAi triggers have a therapeutic window where they are safe but effective.
Materials and Methods
miRNA expression vectors, miR-21 sensor, and rational design. The HBV-miRNA expression cassette consisted of a human U6 promoter, 118 nucleotides of endogenous miR-30 5′ flanking sequence, the pre-mir-30 backbone, and 122 nucleotides of endogenous miR-30 5′ flanking sequence containing a polymerase III (Pol III) termination sequence. HBV-miRs 5, 10, 11, and 12 had Sfold accessibility scores of 7 or 8 on a scale of 8. This basic metric is a probability-weighted sum of four base-pair stacking energies. The weight for a base-pair stack is the probability that the dinucleotide on the target for the stack is single stranded and was estimated by structures predicted by Sfold. This value provides an average accessibility across the whole duplex. The miR Luc and HBV-miR 2 cassettes with flanking sequences were amplified by PCR with primers containing HinDIII and XbaI recognition sites. This fragment was ligated into the HinDIII and XbaI sites of pGL3-Control (Promega) to make pSV40 miR Luc and pSV40 HBV-miR 2. Oligonucleotides with the sequence tcgagTCAACATCAGGACATAAGCTAgc and ggccgcTAGCTTATGTCCTGATGTTGAc were annealed and ligated into the XhoI and NotI sites of Psicheck 2 (Promega) to make the miR-21 Sensor. Bulged nucleotides are shown in boldface.
Tissue culture and RNA isolation. Huh-7 cells were cultured in DMEM supplemented with 10% fetal calf serum or 5% fetal calf serum (at media change after transfection) and 1% each of penicillin/streptomycin (P/S), glutamine, pyruvate, and nonessential amino acids (Invitrogen, Carlsbad, CA). All transfections were carried out in 6-well dishes using Lipofectamine 2000 (Invitrogen). P/S was omitted during transfection. Cells received a total of 4 µg of DNA. In cases where varying amounts of RNAi expression vector was added, the stuffer plasmid pENTR2B (Invitrogen) was added to maintain 4 µg of DNA. For RNAi dose–response studies, 2 µg of the plasmid pTHBV2 [HBV ayw genotype accession #V01460 (ref. 31)] was co-transfected with indicated amounts of RNAi expression plasmid. Cells were cultured for 3 days. In most cases RNAs were purified using Trizol (Invitrogen). In some cases, where only large RNAs were recovered, RNAs were purified using an RNeasy kit (Qiagen, Valencia, CA). For competition experiments against a miRNA, 1.5 µg (Figure 6a) or 0.5 µg (Figure 6b) of Luc miRNA was used, and 0.5 µg of Luc shRNA and 0.07 µg of Luc siRNA were used for shRNA and siRNA competition experiments, respectively. Luc shRNA and Luc siRNA sequences are described in ref. 32. Doses were selected such that similar amounts of silencing for the Luc miRNA, shRNA, and siRNA were observed in the absence of competitor. For Figure 6h, 50 ng of miR-21 reporter was co-transfected with indicated amounts of HBV-miRs 2 or 9. At 3 days, renilla and firefly luciferase were measured using a Dual Luciferase Assay (Promega). Renilla/firefly ratio was calculated and normalized to the signal from cells transfected with a psicheck 2 vector (Promega) that did not contain a miR-21 binding site.
Strand-biasing studies and HBV mRNA northern blots. For strand-biasing studies, cells were transfected with 2 µg of indicated shRNA or HBV-miRNA expression plasmid as well as 2 µg of pTHBV2. RNAs were harvested and 20 µg was resolved by 15% denaturing polyacrylamide gel electrophoresis. RNAs were electrotransferred to Hybond N+ membrane (Amersham Pharmacia Biotech, Piscataway, NJ) and UV-crosslinked. 32P-labeled oligonucleotides complementary to the mature sense or antisense guide strand (Integrated DNA technologies) were hybridized at 37 °C for 16 hours in Church's Buffer (3X SSC, 5 mmol/l Tris-HCl pH 7.4, 0.5% sodium dodecyl sulfate 0.05% Ficol 400, 0.05% polyvinylpyrolidone, and 0.05% bovine serum albumin) and washed for 30 min in 0.5× SSC/0.1% sodium dodecyl sulfate at 37 °C followed by two washes in 0.5× SSC/0.1% sodium dodecyl sulfate. A Decade Marker (Ambion, Austin, TX) was used as a size marker. Bands were detected using a phosphorimager (Molecular Dynamics, Piscataway, NJ). A probe for the U6 spliceosomal RNA was used for normalization. For HBV mRNA northerns, 5 µg of total RNA was treated with NorthernMax-Gly Sample Loading Dye (Ambion) and resolved on a 0.8% Agarose 1× TAE gel. RNAs were transferred to Hybond N+ by capillary transfer and were hybridized to a probe corresponding to the unit-length HBV genome, as above except at 65 °C. The blot was reprobed for glyceraldehyde 3-phosphate dehydrogenase and these values were used to normalize for loading.
Statistical analysis of RNAi potency and competition between miRs and other RNAs. Statistical analysis for Figures 5 and 6 was carried out using SAS version 9.1. For Figure 5a–c, a single-tailed one-way analysis of variance with Dunnet's multiple comparison test was conducted for silencing by triggers that were lower than HBVU6#2. For competition experiments in Figure 6a–e,g,h, a single-tailed one-way analysis of variance with Dunnet's multiple comparison test for silencing levels that were higher than siRNA luc (or miR Luc or shRNA luc depending on data set) was carried out.
Statistical analysis of factors influencing silencing efficiency. The DSSE (in kcal/mol) was calculated as the difference between the four base 5′-antisense stability and the four base 5′-sense stability, i.e., DSSE = AntiS − SS. Internal stability of the antisense siRNA strand was calculated as the sum of four stacking energies for nucleotide 11–14 of the siRNA:target hybrid. ΔGnucleation, ΔGdisruption, and ΔGtotal were calculated as described.25 The entropic penalty for nucleation was estimated to be 4.09 kcal/mol. For the interaction to be productive, ΔGN must be <0.
To account for the large variation in the standard deviations of the measured knockdown levels (Supplementary Table S1), the weighted linear regression method was employed, the weight being the inverse proportion to the standard deviation, so that a data point with smaller standard deviation carries more weight than one with larger standard deviation. In a standard nonweighted regression analysis, R value is the correlation coefficient between a predictor (an energetic parameter here) and the response variable (the measured RNAi knockdown level), and the P value of the regression is the same P value for the significance of the correlation. For weighted regression, the R value was interpreted as a weighted version of the correlation coefficient, and the P value of the regression provides a measure of the significance of the correlation.
Supplementary MaterialFigure S1. Internal stability profile for HBV-miR 1.Figure S2. Internal stability profile for HBV-miR 3.Figure S3. Internal stability profile for HBV-miR 4.Figure S4. Internal stability profile for HBV-miR 5.Figure S5. Internal stability profile for HBV-miR 7.Figure S6. Internal stability profile for HBV-miR 8.Figure S7. Internal stability profile for HBV-miR 9.Figure S8. Internal stability profile for HBV-miR 11.Figure S9. Internal stability profile for HBV-miR 12.Figure S10. Internal stability profile for HBV-miR 13.Figure S11. Internal stability profile for HBV-miR 14.Figure S12. Internal stability profile for HBV-miR 15.Table S1. Energetic parameters and target knockdown level and SD at low dosage and high dosage conditions.
Supplementary Material
Internal stability profile for HBV-miR 1.
Internal stability profile for HBV-miR 3.
Internal stability profile for HBV-miR 4.
Internal stability profile for HBV-miR 5.
Internal stability profile for HBV-miR 7.
Internal stability profile for HBV-miR 8.
Internal stability profile for HBV-miR 9.
Internal stability profile for HBV-miR 11.
Internal stability profile for HBV-miR 12.
Internal stability profile for HBV-miR 13.
Internal stability profile for HBV-miR 14.
Internal stability profile for HBV-miR 15.
Energetic parameters and target knockdown level and SD at low dosage and high dosage conditions.
Acknowledgments
We acknowledge the Computational Molecular Biology and Statistics Core at the Wadsworth Center for providing computing resources. We acknowledge Beverly Davidson, Ryan Boudreau, Ramona McCaffrey, and Thomas Cradick for helpful discussions and critical reading of the manuscript and Michael Henry for reagents. We thank Rebecca Marquez for the gift of the miR-21 reporter. This work was supported in part by US National Science Foundation grants DMS-0200970 (Y.D.) and DBI-0650991 (Y.D.), US National Institutes of Health grants GM068726 (Y.D.), AI068885-01A1 (A.P.M.) and a Center for Gene Therapy Grant DK54759 (A.P.M.).
REFERENCES
- Bruix J., and , Llovet JM.Hepatitis B virus and hepatocellular carcinoma J Hepatol 200339S59–S63.Suppl 1 [DOI] [PubMed] [Google Scholar]
- Perrillo RP. Interferon in the management of chronic hepatitis B. Dig Dis Sci. 1993;38:577–593. doi: 10.1007/BF01316785. [DOI] [PubMed] [Google Scholar]
- Vial T., and , Descotes J. Clinical toxicity of the interferons. Drug Saf. 1994;10:115–150. doi: 10.2165/00002018-199410020-00003. [DOI] [PubMed] [Google Scholar]
- Fattovich G, Giustina G, Favarato S., and , Ruol A. A survey of adverse events in 11,241 patients with chronic viral hepatitis treated with α-interferon. J Hepatol. 1996;24:38–47. doi: 10.1016/s0168-8278(96)80184-x. [DOI] [PubMed] [Google Scholar]
- Dienstag JL, Schiff ER, Wright TL, Perrillo RP, Hann HW, Goodman Z, et al. Lamivudine as initial treatment for chronic hepatitis B in the United States. N Engl J Med. 1999;341:1256–1263. doi: 10.1056/NEJM199910213411702. [DOI] [PubMed] [Google Scholar]
- Meister G., and , Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG., and , Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey AP, Nakai H, Pandey K, Huang Z, Salazar FH, Xu H, et al. Inhibition of hepatitis B virus in mice by RNA interference. Nat Biotechnol. 2003;21:639–644. doi: 10.1038/nbt824. [DOI] [PubMed] [Google Scholar]
- Grimm D., and , Kay MA. Therapeutic short hairpin RNA expression in the liver: viral targets and vectors. Gene Ther. 2006;13:563–575. doi: 10.1038/sj.gt.3302727. [DOI] [PubMed] [Google Scholar]
- Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- ter Brake O, Konstantinova P, Ceylan M., and , Berkhout B. Silencing of HIV-1 with RNA interference: a multiple shRNA approach. Mol Ther. 2006;14:883–892. doi: 10.1016/j.ymthe.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Sen G, Wehrman TS, Myers JW., and , Blau HM. Restriction enzyme-generated siRNA (REGS) vectors and libraries. Nat Genet. 2004;36:183–189. doi: 10.1038/ng1288. [DOI] [PubMed] [Google Scholar]
- Shirane D, Sugao K, Namiki S, Tanabe M, Iino M., and , Hirose K. Enzymatic production of RNAi libraries from cDNAs. Nat Genet. 2004;36:190–196. doi: 10.1038/ng1290. [DOI] [PubMed] [Google Scholar]
- Boden D, Pusch O, Silbermann R, Lee F, Tucker L., and , Ramratnam B. Enhanced gene silencing of HIV-1 specific siRNA using microRNA designed hairpins. Nucleic Acids Res. 2004;32:1154–1158. doi: 10.1093/nar/gkh278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A., and , Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N., and , Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS., and , Khvorova A. Rational siRNA design for RNA interference. Nat Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Ding Y, Chan CY., and , Lawrence CE.Sfold web server for statistical folding and rational design of nucleic acids Nucleic Acids Res 200432W135–W141.Web Server issue [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Chan CY, Maliyekkel A, Lawrence CE, Roninson IB., and , Ding Y. Effect of target secondary structure on RNAi efficiency. RNA. 2007;13:1631–1640. doi: 10.1261/rna.546207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion PL, Salazar FH, Liittschwager K, Bordier BB, Seeger C, Winters MA, et al. A transgenic mouse lineage useful for testing antivirals targeting hepatitis B virus Frontiers in Viral Hepatitis 2003Elsevier Science: Amsterdam; 197In: Schinazi, RF and Sommadossi, JP and Rice, C (eds)209 [Google Scholar]
- Ameres SL, Martinez J., and , Schroeder R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell. 2007;130:101–112. doi: 10.1016/j.cell.2007.04.037. [DOI] [PubMed] [Google Scholar]
- Lu ZJ., and , Mathews DH. Efficient siRNA selection using hybridization thermodynamics. Nucleic Acids Res. 2008;36:640–647. doi: 10.1093/nar/gkm920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long D, Lee R, Williams P, Chan CY, Ambros V., and , Ding Y. Potent effect of target structure on microRNA function. Nat Struct Mol Biol. 2007;14:287–294. doi: 10.1038/nsmb1226. [DOI] [PubMed] [Google Scholar]
- Castanotto D, Sakurai K, Lingeman R, Li H, Shively L, Aagaard L, et al. Combinatorial delivery of small interfering RNAs reduces RNAi efficacy by selective incorporation into RISC. Nucleic Acids Res. 2007;35:5154–5164. doi: 10.1093/nar/gkm543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holen T, Amarzguioui M, Babaie E., and , Prydz H. Similar behaviour of single-strand and double-strand siRNAs suggests they act through a common RNAi pathway. Nucleic Acids Res. 2003;31:2401–2407. doi: 10.1093/nar/gkg338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus MT, Haines BB, Dillon CP, Whitehurst CE, van Parijs L, Chen J, et al. Small interfering RNA-mediated gene silencing in T lymphocytes. J Immunol. 2002;169:5754–5760. doi: 10.4049/jimmunol.169.10.5754. [DOI] [PubMed] [Google Scholar]
- Koller E, Propp S, Murray H, Lima W, Bhat B, Prakash TP, et al. Competition for RISC binding predicts in vitro potency of siRNA. Nucleic Acids Res. 2006;34:4467–4476. doi: 10.1093/nar/gkl589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ, Mello CC., and , Zamore PD. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2:E98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galibert F, Mandart E, Fitoussi F, Tiollais P., and , Charnay P. Nucleotide sequence of the hepatitis B virus genome (subtype ayw) cloned in E. coli. Nature. 1979;281:646–650. doi: 10.1038/281646a0. [DOI] [PubMed] [Google Scholar]
- McCaffrey AP, Meuse L, Pham TT, Conklin DS, Hannon GJ., and , Kay MA. RNA interference in adult mice. Nature. 2002;418:38–39. doi: 10.1038/418038a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Internal stability profile for HBV-miR 1.
Internal stability profile for HBV-miR 3.
Internal stability profile for HBV-miR 4.
Internal stability profile for HBV-miR 5.
Internal stability profile for HBV-miR 7.
Internal stability profile for HBV-miR 8.
Internal stability profile for HBV-miR 9.
Internal stability profile for HBV-miR 11.
Internal stability profile for HBV-miR 12.
Internal stability profile for HBV-miR 13.
Internal stability profile for HBV-miR 14.
Internal stability profile for HBV-miR 15.
Energetic parameters and target knockdown level and SD at low dosage and high dosage conditions.