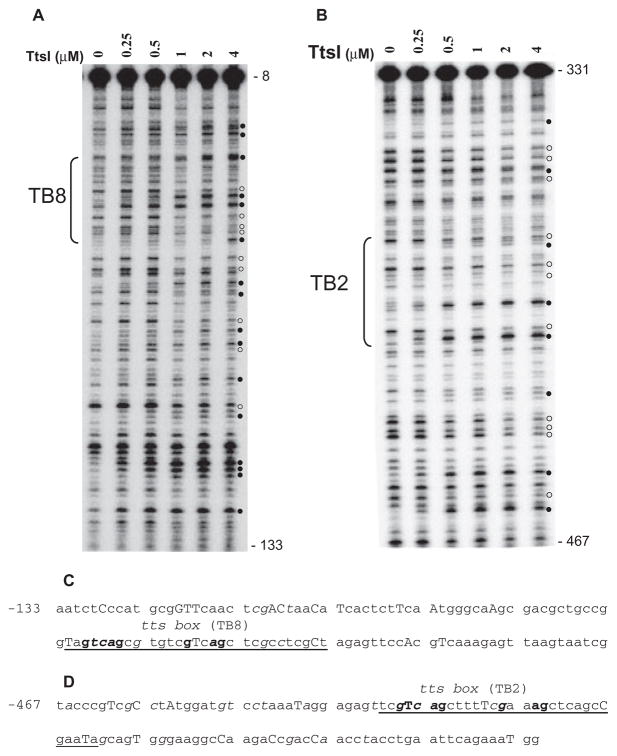
Fig. 4.
DNase I footprints of TtsI bound to the nopB (TB8) and rmlB (TB2) promoter regions. 32P-labelled PCR fragments were incubated with different concentrations of TtsI and digested using limiting concentrations of DNase I. After purification, DNA fragments were separated in a 6% sequencing gel, dried, exposed to Phosphoimager screens and scanned. The TB8- and TB2-containing PCR fragments are shown in A and B respectively. Numbers on the right refer to the DNA position compared with ATG of the downstream gene. Brackets indicate the limits of the TB. Open and closed circles pinpoint DNase I protected and hyperreactive bands respectively. Below the panels, the DNA sequences show the organization of the nopB (C) and rmlB (D) promoter regions and their reactivity to DNase I cleavage. Underlined bases delineate the TBs with the highly conserved bases in bold font. Capital letters represent DNase I hyperreactive bases and italics show protected bases. Numbers refer to positions upstream of the ATG of the downstream gene.