Figure 2.
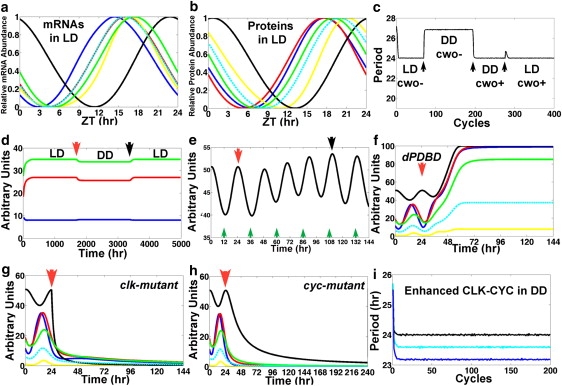
The model replicates biological results. (a) Plot of the results of model simulations in LD of the concentrations of clk (black; peak, 22.8 h), per (blue; peak, 14.4 h), tim (red; peak, 16.1 h), cwo (yellow; peak, 16.8 h), Pdp1 (green; peak, 17.4 h), and vri (cyan; peak, 15.9 h). (b) Plot of the results of model simulations in LD of the concentration of the CLK (black; peak, 0.7 h), PER (blue; peak, 17.7 h), TIM (red; peak, 17.6 h), CWO (yellow; peak, 21.2 h), PDP1 (green; peak, 19 h), and VRI (cyan; peak, 20 h). Relative abundance is determined by (x − min)/(max − min). The Zeitgeber reference time is described in LD conditions only. To define a time reference in DD conditions we set time 0 at −0.7 h of the peak of the CLK protein so that the latter peaks at the same time in both LD and DD. The peak times of the direct-target genes and proteins in DD are as given in a except for TIM, which peaks at 16.6 h. (c) Plot of the period (time between the peaks of per mRNA) in an experiment where the model cycles first in LD with cwo mutation (cwo−), then in DD with cwo mutation, then in DD with wt cwo (cwo+), and finally in LD with wt cwo. Transitions are applied at ZT = 0 (arrows). Notice the minimum variability of the 24-h period in both DD and LD in wt cwo simulations. (d) Plot of the peak (green) and trough (blue) levels and the amplitude (peak − trough, red) of per mRNA, computed from an experiment where the model cycles first in LD (0 to red arrow), then in DD (red arrow to black arrow), and finally in LD. Transitions from LD to DD and from DD to LD are applied at ZT = 0. (e) The model clock resets (black arrow) 4.5 days after a shift of +12 h is applied at the red arrow. Notice that CLK evolves to a peak around midnight of the previous time zone (black arrow). Green arrows point to ZT = 12 h of the previous time zone. (f–h) Plots of the simulated dynamics of dPDBD (in DD) (f), clk (in LD) (g), and cyc (in DD) (h) null mutations applied at time = 24 h (red arrows). Notice the elevated and constant levels of per (blue) and tim (red) mRNAs in simulated dPDBD mutants and the low levels of per and tim mRNAs in clk- and cyc-null mutants. The mRNA levels of per, tim, cwo, Pdp1, and vri are shown in blue, red, yellow, green, and cyan, respectively. The CLK protein is shown in black. (i) Plot of the period of the wt model (black) and the simulations where all the positive regulatory weights of CLK-CYC on direct-target genes are increased by 15% (cyan) or 30% (blue) in DD conditions.
