Figure 7.
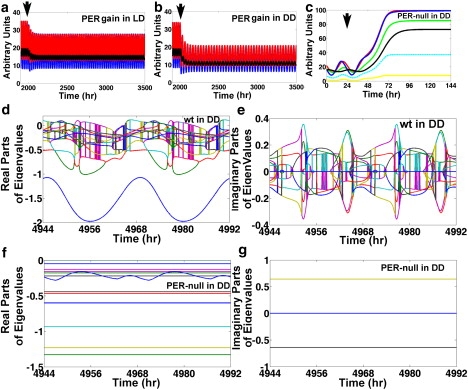
Simulations of PER gain and loss of function. (a and b) Reactions of per (blue), tim (red), and clk (black) mRNAs when λper, PER is increased by 25% (PER gain of function) at the arrows in LD and DD, respectively. (c) Results of a simulation where a per-null mutation (λper, PER = 0) is applied at the arrow for per (blue), tim (red), clk (black), cwo (yellow), vri (cyan), and Pdp1 (green) direct-target genes. (d and e) Real and imaginary parts, respectively, of the eigenvalues (colored lines) of the flow matrix of the wt model in DD; observe that the real parts cross the x axis. (f and g) Real and imaginary parts of the eigenvalues (colored lines) of the flow matrix of the per-null model in DD; all the real parts are negative and only two conjugate imaginary parts are nonzero. Notice that times 4944 h, 4968 h, and 4992 h correspond to ZT = 0 h, and times 4956 h and 4980 h correspond to ZT = 12 h.
