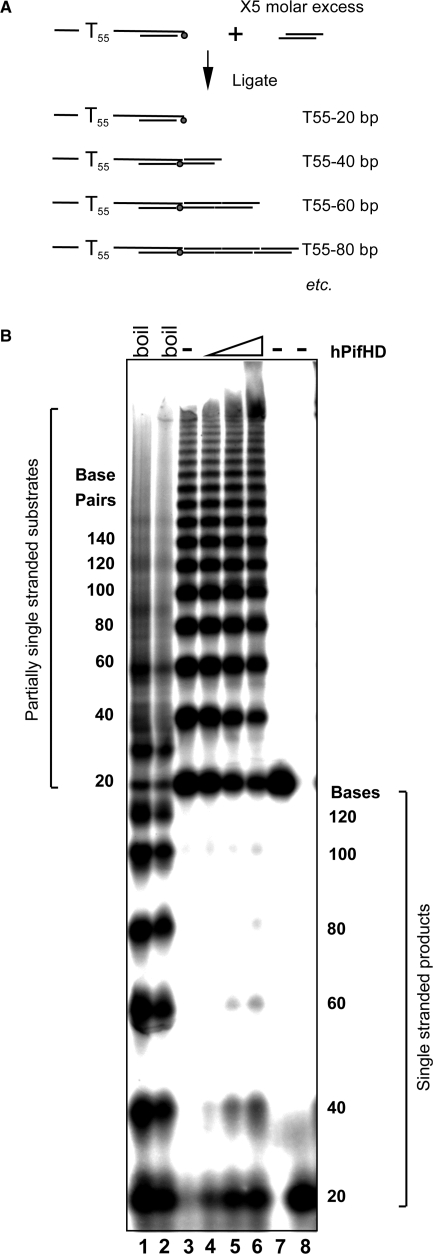
Figure 6.
Action of hPifHD on substrates with an extended duplex. The ability of hPifHD to unwind progressively longer stretches of duplex DNA was assessed using a population of substrate molecules with a 55-base 5′ T tail and a duplex portion increasing in 20 base increments. (A) Schematic diagram of substrate generation. Two pairs of phosphorylated oligonucleotides were annealed as illustrated. The oligonucleotide annealed to the strand with a 55 base T extension was phosphorylated with (γ-32P)ATP. Double stranded oligonucleotides were ligated with a 5-fold molar excess of the non-tailed duplex oligo generating a series of products with a 55 base 5′ T tail and duplex portions increasing in 20 bp increments. (B) Unwinding products were resolved on a 6% (19 : 1) native poly-acrylamide gel. Lanes 1 and 2 show the substrates heat-denatured with and without formamide present. Lane 3 is the native substrate and lanes 4–6 unwinding product generated by hPifHD (2.5, 10 and 40 nM hPifHD). Lanes 7 and 8 are markers for the minimal substrate (55 bases 5′ ssDNA, 20 bp duplex), native and denatured respectively. The fractions (%) unwound for the 20, 40, 60 and 80 bp substrate molecules were 18, 8, 1.3 and 0% at 2.5 nM hPifHD; 41, 27, 8.5 and 1.8% at 10 nM hPifHD and 47, 43, 12.4 and 5% at 40 nM hPifHD.