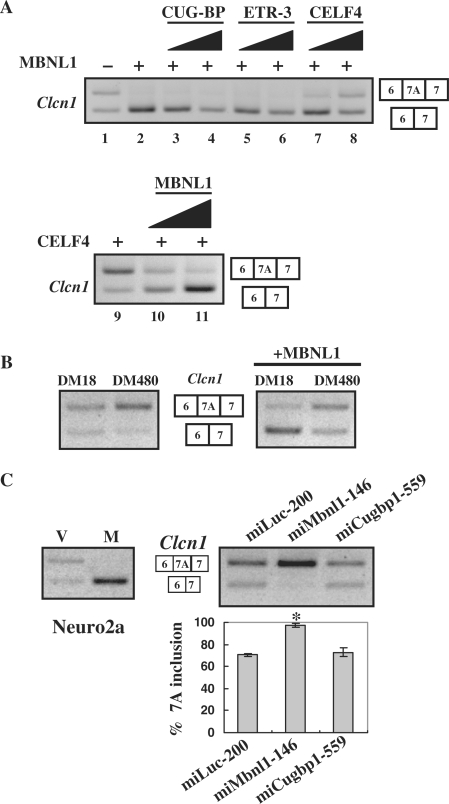
Figure 2.
MBNL1 is antagonized by CELF4 and expanded CUG repeats. (A) Antagonistic effects of CELF4 against MBNL1 in the splicing regulation of Clcn1. Clcn1 minigene was co-transfected with MBNL1 or MBNL1 plus CELF protein into COS-7 cells. Upper panel: lane 1, the pattern of empty vector-transfected cells; lanes 2–8, a constant amount of MBNL1-encoding vector and increasing amounts of one of the CELF-encoding vectors were co-transfected at ratios of 1 : 0 (lane 2), 1 : 1 (lanes 3, 5 and 7) and 1:3 (lanes 4, 6 and 8). Lower panel: similarly, a CELF4-encoding vector and MBNL1-encoding vector were transfected at ratios of 1 : 0 (lane 9), 1 : 1 (lane 10) and 1 : 3 (lane 11). The total amount of transfected plasmids was held constant by adjusting the amounts of empty vector. (B) Effects of an expanded CUG repeat on the chloride channel splicing. Clcn1 minigene was transfected with an expression vector of DMPK harboring a normal (DM18) or expanded (DM480) CTG repeat (left panel). Either normal or expanded DMPK vector was co-transfected with MBNL1 and the Clcn1 minigene, and the splicing patterns were analyzed (right panel). (C) Results of Clcn1-splicing assays in Neuro2a cells. MBNL1-mediated repression of exon 7A inclusion in Neuro2a cells (left). ‘V’ and ‘M’ indicate empty vector and MBNL1, respectively. Note that the basal inclusion of exon 7A in Neuro2a cells was higher than that in COS-7 cells. Clcn1-splicing regulation was affected by RNAi of Mbnl1 but not Cugbp1 in Neuro2a cells (right). Bar chart shows the quantified results of exon 7A inclusion (mean ±SD, n = 4). According to ANOVA and Dunnett's test, miMbnl1-146 induced a statistically significant increase of exon 7A compared to miLuc-200 (*P < 0.0001), whereas miCugbp1-559 did not (P = 0.36).