Abstract
The tight regulation of transiently expressed antimicrobial peptides (AMPs) with a distinct antimicrobial spectrum and different expression kinetics contributes greatly to the properly regulated immune response for resistance to pathogens and for the maintenance of mutualistic microbiota in Drosophila. The important role of differential regulation of AMP expression at the posttranscriptional level needs to be elucidated. It was observed that the highly expressed Cecropin A1 (CecA1) mRNA encoding a broad antimicrobial spectrum AMP against both bacteria and fungi decayed more quickly than did the moderately expressed Diptericin mRNA encoding AMP against Gram negative bacteria. The mRNA stability of AMPs is differentially regulated and is attributed to the specific interaction between cis-acting ARE in 3′-UTR of AMP mRNA and the RNA destabilizing protein transactor Tis11 as shown in co-immunoprecipitation of the Tis11 RNP complex with CecA1 mRNA but not other AMP mRNA. The p38MAPK was further demonstrated to play a crucial role in stabilizing ARE-bearing mRNAs by inhibiting Tis11-mediated degradation in LPS induced AMP expression. This evidence suggests an evolutionarily conserved and functionally important molecular basis for and effective approach to exact control of AMP gene expression. These mechanisms thereby orchestrate a well balanced and dynamic antimicrobial spectrum of innate immunity to resist infection and maintain resident microbiota properly.
INTRODUCTION
The innate immune system is critical for the host not only to control microbial infection at the front line of immune defense, but also to maintain the mutualistic relationship with the resident microbiota community, which is of great importance for supporting and sustaining health. An essential aspect of the Drosophila melanogaster immune response, which is equivalent to innate immunity in mammals, is the spatially and temporally regulated expression of a battery of antimicrobial peptides (AMPs) (1–5). The AMPs have different spectra of activity, targeting different classes of pathogenic microorganisms (1–3). Diptericin (Dpt), Drosocin (Dro) and Attacin (Att) act against Gram-negative bacteria. Defensin (Def) is active against Gram-positive bacteria, whereas Drosomycin (Drs) and Metchnikowin (Mtk) are antifungal agents. Cecropin A1 (CecA1) has a broad antibacterial spectrum against both bacteria and fungi (6,7). Each AMP works in concert with others to take effect with an integrated and proper antimicrobial scope. Because of the distinct antibacterial spectrum of each AMP, the tight regulation of extent and duration of individual AMP expression contributes greatly to the overall effect on the resistance to pathogens and the maintenance of the resident microbiota (8,9). Therefore, it is important to elucidate the differentially regulated gene expression of AMPs with distinct kinetics and antimicrobial spectra at multiple levels. Such an understanding would lead to the revelation of its crucial role in the orchestration of effective and efficient antimicrobial spectra by precise expression control of distinct AMPs in properly regulated host immune responses.
Although AMPs are constitutively synthesized in specific tissues at a basal level (3), a characteristic aspect of AMP synthesis in D. melanogaster is the transient expression of a battery of antimicrobial peptides upon immune response, which is critical for protection against many microbial pathogens (1–3,10,11). Prior to infection, most AMP mRNA levels are very low, but transcripts accumulate rapidly after infection (3,5,12). From then on, AMP mRNA levels decrease (3,12). Certainly, the transient expression of AMP genes is tightly regulated so that insects can response to antigen quickly to resist the predation of rapidly dividing pathogenic microorganisms, and then withdraw highly active AMPs successively to avoid prolonged inhibition of mutualistic microbiota in the host (8–11). This delicate regulation depends upon the interplay among elements that control gene expression at multiple levels such as transcription, mRNA stability and translation (13).
Undoubtedly, transcriptional control at κB-like sites bound by Rel family proteins (14–18) is a determinant of the distinct spectra and of the stereotypical kinetics of AMP gene expression activated by different microbes through Toll and IMD pathways (3,5,11,19). In fact, in addition to transcription, posttranscriptional events, particularly the stability of specific mRNA, are also important determinants of the extent and duration of gene expression (12,20–23). After analysis of sequence motifs in Drosophila AMPs, it was observed that the mRNA 3′-UTR of quite a few AMPs contain AU-rich sequences similar to the adenylate and uridylate rich element (AU-rich element, ARE). This is a highly conserved posttranscriptional regulatory element found throughout evolution from yeasts and insects to mammals (24–26). In mammalian cells, ARE controls mRNA stability via interactions with specific RNA binding proteins: some ARE-binding proteins (AUBPs) target the transcript for degradation, such as tristetraprolin (TTP), whereas others, such as HuR, mediate transcript stabilization (20,26–29). In addition, it has been implied that AREs can exert either a stabilizing or destabilizing effect on mRNA depending upon the p38 mitogen-activated protein kinase (p38MAPK) activity within the mammalian cells (30,31). In general, the ARE sequences specifically bound by AUBPs include a central AU3-5A core with a UU contributed from the AU3-5A on either side and are always found in the 3′-UTR of a variety of immediate response genes, including those encoding cytokines, inflammatory mediators and other such molecules (25,26,28,32,33). In light of the presence of ARE motifs in the 3′-UTR of AMP mRNA, one may speculate that the transient expression of AMPs may be under tight regulation at the posttranscriptional level, particularly with regard to transcript stability control. Thus, it is important to explore the fundamental aspects of and relevant approaches to mRNA stability regulation that control AMP expression differentially, and that could ultimately translate the distinct expression kinetics of each AMP gene into a dynamic and delicately orchestrated antimicrobial spectrum combining each AMP’s intensity, duration and specific activity.
In this study, posttranscriptional analyses of gene expression of representative AMPs including CecA1 and Dpt, which have similar but not identical ARE sequences located in the 3′-UTR, were performed to elucidate the differential regulation of mRNA stability in AMP gene expression and the crucial role of p38MAPK in stabilizing ARE-bearing mRNAs by inhibiting Tis11-mediated degradation in LPS induced AMP expression in Drosophila macrophage-like S2* cells.
MATERIALS AND METHODS
Reagents
Escherichia coli Lipopolysaccharides (LPS) (62326), SB203580 and 20-hydroxy-ecdysone (H5142) were obtained from Sigma-Aldrich. Actinomycin D (Act.D) was purchased from Ameresco, His-tag antibody from Abmart. The gene-specific primers were synthesized by Invitrogen China.
Cell culture
Drosophila Schneider (S2*) cells (kindly provided by Dr Ge Baoxue) were cultured in 1×Schneider’s Drosophila medium (Invitrogen) supplemented with 10% FBS (Invitrogen), 50 U/ml penicillin and 50 ng/ml streptomycin at 25°C (34). For the treatment with LPS in which peptidoglycan (PGN) was the immune activator with the ability to activate the IMD pathway (35,36), S2* cells were incubated with 1 µM 20-hydroxy-ecdysone to induce differentiation for at least 24 h prior to stimulation with 10 µg/ml LPS. For p38MAPK inhibition, cells were incubated with 10 μM SB203580 or with vehicle (1% dimethyl sulfoxide, DMSO) for 30 min prior to stimulation.
Plasmid constructs
The firefly luciferase (Fluc) gene and polyA signal of SV40 from pGL-3 Basic (Promega) was sub-cloned into pAc5.1-Flag-V5-His C vector with KpnІ and SalІ to construct pAC-Fluc plasmid. Using the S2* cells cDNA as templates, the 3′-UTR of Dpt (147 bp), heat shock protein 70 (HSP70) (234 bp) and ribosomal protein 49 (rp49) (162 bp), and fragments of I (full length of CecA1 3′-UTR, 97 bp), II (70 bp), III (55 bp), IV (42 bp), V (19 bp) of CecA1 3′-UTR were synthesized by PCR with specific primers (Supplementary Table S1). The 3′-UTR of TNF-α was PCR amplified using THP1 cells cDNA as templates. Excised with XbaІ or NheІ and purified with a Gel Extract kit (Omega). DNA fragments were ligated into the unique XbaІ site of the pAC-Fluc, located downstream of the Fluc coding sequence. The renilla luciferase (Rluc) gene from pRL-SV40 (Promega) was ligated into NheІ-XbaІ sites of pAC-Fluc to replaced Fluc for pAC-Rluc plasmid construction. The coding region of Drosophila Tis11 was PCR amplified and sub-cloned into pAc5.1-Flag-V5-His C vector with the KpnІ and NotІ sites residing downstream of actin5C promoter and upstream of 6×His tag to make pAC-Tis11-His. The coding region of human TTP from pCDNA3.0-myc-TTP plasmid was sub-cloned into pAc5.1-Flag-V5-His C plasmid with HindШ and NotІ to construct pAC-TTP plasmid. All DNA constructs were verified by DNA sequencing (Invitrogen, Shanghai, China).
RNA isolation and analysis of gene expression
Total RNA was isolated from S2* cells using Trizol reagent (Invitrogen) and treated with DNase (Promega). Total RNA (1 µg) was used together with MMLV reverse transcriptase (Promega) and oligo (dT)18 primer to synthesize first strand cDNA, which was used as a template for quantitative real time RT–PCR (qRT–PCR) with gene-specific primer pairs (Supplementary Table S2) and SYBR Green PCR master mix (Toyobo) on an ABI PRISM 7900 Fast Real Time PCR System (Applied Biosystems). The expression level of CecA1 or Dpt was normalized to rp49 and Fluc was normalized to Rluc in each sample in order to quantify the relative levels of a given mRNA according to the ΔCt analysis. For absolute quantitative real time RT–PCR, the DNA standard samples for AMPs and rp49 were made by PCR amplification and quantified using a spectrophotometer (Beckman DU800). Absolute quantitative real time RT–PCR was performed using serially diluted standard samples as templates to make a standard amplification curve.
Act.D chase studies for mRNA stability measurement
For mRNA stability assays, S2* cells were incubated with 10 µg/ml Act.D to inhibit transcription. At the indicated time points after the addition of Act.D, cells were harvested and total RNA was extracted. The expression levels of CecA1, Dpt and Fluc at each time point were measured by qRT–PCR as described earlier and normalized to the according rp49 levels. The remaining mRNA was determined by comparison with the expression level of the relevant gene at the zero time point (designated 100%) when Act.D was added.
Cell transfection and luciferase reporter assay
S2* cells were seeded at a density of 1 × 106 cells/ml in 6-well plates. The next day, 0.5 µg Fluc reporter plasmids containing the indicated fragment from 3′-UTR of various genes and 0.5 µg Rluc reporter plasmids (pAC-Rluc) were co-transfected into cells using the calcium phosphate precipitation method (Invitrogen). Three days after transfection, cells were harvested and luciferase activities were measured according to the recommended procedures for the dual luciferase assay system (Promega) on a PerkinElmer Lumat LB 9507 luminometer. The Fluc activity was normalized to the activity of Rluc.
RNA interference
The primer sequences (Supplementary Table S3) used to generate templates for synthesizing double-strand RNA (dsRNA) of Tis11 and EGFP included a 5′ T7 RNA polymerase-binding site. For Tis11 dsRNA template amplification, S2* cells cDNA was used as template in RT–PCR, while pEGFP-C1 vector (Clontech) was used as template to amplify EGFP dsRNA template. The purified PCR products were used as templates to produce dsRNA by using a MEGAscript RNAi kit (Ambion). A 15 µg dsRNA was transfected into 2 ml S2* cells (1 × 106/ml in 6-well plate) using a calcium phosphate precipitation method (see above) with or without reporter plasmid.
Immunoprecipitation and complex analysis
S2* cells were seeded in 6-well plates and transiently transfected with pAC-Tis11-His plasmid or blank control pAc5.1-Flag-V5-His C vector using a calcium phosphate precipitation method (see above). Three days later, the S2* cells were stimulated with LPS for 3 h, inducing AMP mRNA to high levels. The cells were then lysed for 10 min on ice in RNA immunoprecipitation (RIP) buffer [50 mM Tris (pH 7.4), 150 mM NaCl, 0.5% Nonidet P-40, 1 mM DTT, 100 U/ml RNase inhibitor (Promega), 2.5% proteinase inhibitor cocktails (Sigma), 2 mM vanadyl ribonucleoside complexes (NEB)]. The cell lysate was centrifuged at 14 000 g for 10 min at 4°C and the supernatant of the cytoplasmic lysate was collected for RNA IP assays. Precleared with protein-A Sepharose beads (Amersham), lysates were incubated with 1 µg mouse His-tag antibody to immunoprecipitate Tis11-His fusion protein and rotated for 12 h at 4°C. Five percent (v/v) protein-A beads were added for another 4 h and were washed several times with RIP buffer for IP complex isolation. RNA was extracted from the IP complex and the presence of specific mRNAs in the IP complex was determined by RT–PCR with gene-specific primers. A low-level signal of housekeeping transcript rp49 was detectable in all samples and served to monitor the quality and evenness of sample input.
RESULTS
Transiently expressed AMP genes have distinct kinetics and show substantial differences in mRNA stability
Posttranscriptional regulatory studies of the expression of genes encoding AMPs with distinct antimicrobial spectra were performed on the widely used Drosophila cellular model—macrophage-like S2 cells which can express a spectrum of AMPs in response to immune stimulants and can thus allow more precise manipulation.
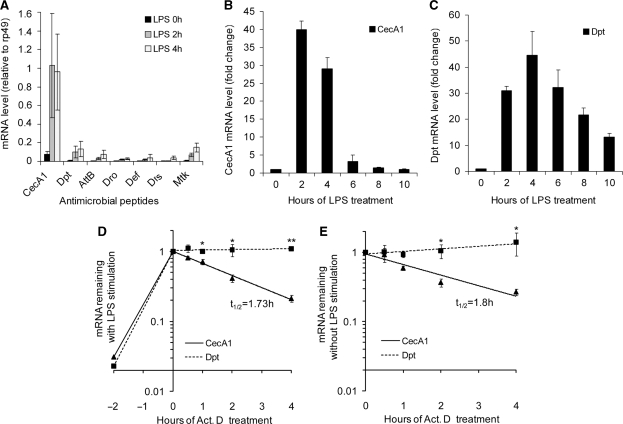
To determine the similarity and dissimilarity in expression profiles of AMP genes, an absolute quantitative real-time RT–PCR method was used to compare the mRNA expression levels of different AMPs. As shown in Figure 1A, whether or not S2* cells were stimulated with LPS for 2 and 4 h, the mRNA of CecA1 was the most abundant transcript among the detected AMPs including Dpt, Def, Dro, AttB, Mtk and Drs. The mRNA levels of the other AMPs were moderate, while Dpt mRNA displayed a relatively high level. To investigate whether the gene expression of the CecA1 and other AMPs were differentially regulated, we first examined the expression kinetics of CecA1 and Dpt by comparing their mRNA time course after immune stimulation. Figure 1B and C show the CecA1 and Dpt mRNA expression profiles from 0 to 10 h in S2* cells stimulated with LPS. Both had similar fold changes of mRNA levels when stimulated and showed comparable transient expression characteristics. However, it was observed that the CecA1 mRNA decreased rapidly to almost basal level within 4–6 h, whereas the Dpt mRNA remained at a high level for up to 10 h. Obviously, the two AMP mRNAs had different expression kinetics as a result of differential regulation.
Figure 1.
The gene expression profiles and mRNA stability of different AMPs. (A) Absolute quantitative real-time RT–PCR analysis of the amount of AMPs and rp49 transcript in S2*cells treated with 10 µg/ml LPS for 0, 2 and 4 h, with the use of an amplification standard curve for each gene. Taking the amount of rp49 as 1, the expression level of each AMP was calculated. (B and C) The qRT–PCR analysis of CecA1 (B) and Dpt (C) mRNA in S2* cells treated with 10 µg/ml LPS for the time indicated. The fold changes of CecA1 and Dpt mRNA were detected at each time point, taking the mRNA level at 0 h as 1. (D and E) S2* cells were stimulated with LPS for 2 h (D) or not stimulated (E), and further incubated with 10 µg/ml Act.D for the time indicated. The qRT–PCR was performed to detect CecA1 and Dpt mRNA remaining at each time point, taking the mRNA level at the time of Act.D added as 1. Values represent the mean ± SD (n = 3 independent experiments), *P<0.05; **P<0.01 for CecA1 versus Dpt at each time point.
To determine whether mRNA stability was involved in the difference between the two AMP expression profiles, Act.D chase studies were used to eliminate effects of transcription on AMP expression and to determine the rate of mRNA decay. As shown in Figure 1D, the decay rate of CecA1 mRNA (t1/2 = 1.73 h) was much quicker than that of Dpt mRNA which remained stable (t1/2 > 4 h) throughout the course of the 4-h observation with LPS treatment (Figure 1D). The results clearly show that CecA1 mRNA is much more unstable than Dpt mRNA, indicating that mRNA stability is an important determinant in the differences in extent and duration of AMP gene expression after immune stimulation.
To test whether the stability of AMP mRNA was influenced by immune stimulation, CecA1 and Dpt mRNA decay rates were also detected in S2* cells at basal level without LPS treatment (Figure 1E). Nevertheless, the decay rate of CecA1 mRNA was almost the same as the stimulated one with a half-life of 1.8 h, while Dpt mRNA was significantly stable during the 4-h observation period. Thus, the mRNA stability of each AMP was not altered whether or not S2* cells were stimulated with LPS (Figure 1, compare D and E). These results suggest that the decay rates of CecA1 and Dpt mRNAs were well maintained in a constitutive manner and that the mRNA stability of both AMPs were under posttranscriptional regulatory control which could sustain a steady decay rate for the transcript in response to LPS stimulation.
In contrast to Dpt 3′-UTR, the CecA1 3′-UTR is sufficient to confer instability on a reporter mRNA
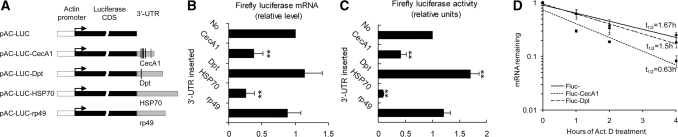
As for the posttranscriptional control of mRNA stability, the AU-rich sequences can be found in the 3′-UTR of several AMP mRNAs. Both the CecA1 3′-UTR and Dpt 3′-UTR contain AU-rich sequences which may act as cis-acting elements in posttranscriptional regulation. Therefore, reporter gene constructs of the luciferase reporter assay system were created by inserting DNA encoding the 3′-UTR of CecA1 or Dpt downstream of the Fluc gene which was driven by actin5C promoter (Figure 2A). To validate the reliability of this method, we used the 3′-UTR of rp49, a housekeeping gene, as a stable control and HSP 70 3′-UTR as an unstable control (37) for the reporter system. Insertion of the HSP70 3′-UTR resulted in great reduction of Fluc at protein level and mRNA level with up to 80% expression inhibited, whereas insertion of rp49 3′-UTR had a little but not significant effect on the expression as compared with pAC-Fluc control vector without 3′-UTR sequence inserted (Figure 2B and C). Subsequently, the regulatory roles of AMPs 3′-UTR were analyzed using this effective and efficient reporter system for posttranscriptional studies.
Figure 2.
CecA1 and Dpt 3′-UTR influenced reporter expression differently. (A) Diagram of reporter gene constructs pAC-Fluc-3′-UTR. The 3′-UTR of CecA1, Dpt, rp49 and HSP70 (sequences shown in Supplementary Table 4) were inserted downstream of the reporter Fluc gene coding region which is indicated by the black bar. The 3′-UTR is represented by the shaded bar of proportional length. Thin lines indicate the AU-rich sequence sites in CecA1 and Dpt. (B) Fluc mRNA levels in S2* cells co-transfected with Fluc reporter plasmids containing the 3′-UTR of different genes and Rluc reporter plasmid as normalization control. The qRT–PCR was performed to detect the transcript of Fluc. Fluc mRNA measured in cells transfected with the pAC-Fluc control vector was designated as 1. Values represent the mean ± SD of at least four experiments (**P < 0.01 for each construct versus pAC-Fluc). (C) Luciferase activity assays of S2* cells co-transfected with Fluc reporter plasmids containing the 3′-UTR of different genes and Rluc reporter plasmid used as normalization control. Fluc activity measured in cells transfected with the pAC-Fluc control vector is designated as 1. Values represent the mean ± SD of at least four experiments (**P < 0.01 for each construct versus pAC-Fluc). (D) S2* cells were transfected with Fluc reporter plasmids containing the 3′-UTR of CecA1, Dpt or no 3′-UTR inserted, and then incubated with Act.D for the time indicated. The qRT–PCR was performed to detect Fluc mRNA remaining at each time point, taking the Fluc mRNA level of each construct at the time of Act.D addition as 1. Values represent the mean ± SD (n = 3 independent experiments).
Compared with the control in which no 3′-UTR was inserted, inserting the 3′-UTR of CecA1 downstream of the Fluc resulted in a 60% decrease in Fluc expression at both mRNA level and protein level (Figure 2B and C). This indicated that CecA1 3′-UTR control of reporter expression mainly occurred at the mRNA level and that the CecA1 3′-UTR harbors a cis element necessary to destabilize the reporter Fluc mRNA. In contrast, inserting the Dpt 3′-UTR increased Fluc activity up to 70% but showed no significant changes at Fluc mRNA level, implying that a cis element modulating translation efficiency could exists within the Dpt 3′-UTR.
To further examine whether the change of Fluc mRNA level under the control of different AMP 3′-UTR might be due to mRNA stability, Act.D chase studies were used. As shown in Figure 2D, the Fluc mRNA decay rate observed with CecA1 3′-UTR (t1/2 = 0.63 h) was faster than that seen with Dpt 3′-UTR (t1/2 = 1.5 h) or with no insert (t1/2 = 1.67 h).
These results demonstrate that the 3′-UTR of different AMPs contributes greatly to the mRNA stability which in turn affects gene expression differentially.
The ARE in the proximal region of CecA1 3′-UTR is the cis-acting element destabilizing mRNA
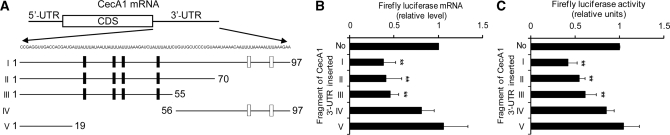
While the AU-rich sequence in Dpt mRNA 3′-UTR has been characterized for its binding ability with AUBP in a previous study (38), the CecA1 mRNA 3′-UTR contains several AU-rich sequence motifs which are scattered in the proximal region and the distal region. To clarify the role of these AU-rich sequences and to localize the cis element of CecA1 3′-UTR contributing to downregulation of gene expression, a series of fragments of CecA1 3′-UTR was subcloned into the 3′-end of the Fluc coding sequence to generate various Fluc reporter constructs (Figure 3A). In comparison with control vector with no insert, fragments II and III both containing the AU-rich sequence in the proximal region decreased Fluc mRNA level significantly and resembled the inhibition effect of the full 3′-UTR (fragment I) to a great extent (Figure 3B). Fragment IV, containing the AU-rich sequence in the distal region, only had a weak effect without significance in reducing Fluc mRNA level. Fragment V, the short proximal region excluding AU-rich sequence, had no obvious effect. Furthermore, as shown in Figure 3C, the Fluc protein activity influenced by different fragments of CecA1 3′-UTR was consistent with its own mRNA level, demonstrating that the suppression of Fluc activity is due to reduction at the mRNA level. Overall, these results indicate that the AU-rich sequence (nt 20 to 55) in the proximal region is the AU-rich element (ARE) contributing to the decreased reporter expression and that this CecA1 ARE in 3′-UTR can control its mRNA stability and regulate CecA1 expression.
Figure 3.
Effects of CecA1 3′-UTR fragments on reporter expression. (A) The sequence of CecA1 3′-UTR and schematic of the different fragments inserted downstream of the Fluc. AUUUA pentamer or AUUUUA hexamer are underlined. Each rectangle indicates an AU-rich sequence site. The AU-rich sequence with U surrounding it is indicated as a black rectangle. (B) Fluc mRNA levels in S2* cells which were co-transfected with Fluc reporter plasmids containing the different fragments of CecA1 3′-UTR and Rluc reporter plasmid as normalization control. The qRT–PCR was performed to detect the expression level of Fluc mRNA. Fluc mRNA measured in cells transfected with the pAC-Fluc control vector is designated as 1. Values represent the mean ± SD of at least four experiments (**P < 0.01 for each construct versus pAC-Fluc). (C) Luciferase activity assays of S2* cells co-transfected with Fluc reporter plasmids that were inserted with different fragments of CecA1 3′-UTR and Rluc reporter plasmid used as normalization control. Fluc activity measured in cells transfected with the pAC-Fluc control vector is designated as 1. Values represent the mean ± SD of at least four experiments (**P < 0.01 for each construct versus pAC-Fluc).
Identification of Tis11 as a transactor specifically regulating CecA1 expression through ARE in the 3′-UTR
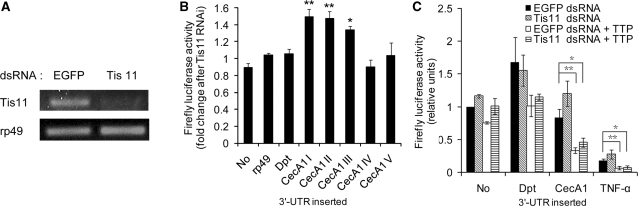
AREs exert their effect on gene expression through interaction with specific ARE binding proteins (AUBPs) or with the help of microRNA (miRNA) (20–23,26,39–41). To identify genes that are required for CecA1 mRNA instability control, a list of dsRNAs that target the known AUBPs, including Tis11, ELAV, RBP9 and small RNA processing factor Dicer1 were generated and used in the AUBPs knockdown by RNA interference (RNAi). These dsRNAs were co-transfected into S2* cells with a reporter construct containing the indicated 3′-UTR and Fluc activity changes were measured. Compared with the Fluc control construct with no 3′-UTR inserted, the treatments of dsRNA specific for ELAV, RBP9 or Dicer1 had no obvious effect on Fluc activity for constructs inserted with different 3′-UTR (data not shown). However, dsRNA-treatments targeting Tis11 (Figure 4A), a homologue of mammalian TTP, increased Fluc activity ∼50% for the reporter construct inserted with CecA1 3′-UTR (Figure 4B). This effect appeared specifically mediated by the 3′-UTR of CecA1 since knocking down Tis11 by RNAi had no obvious effect on Fluc activity changes when the constructs were inserted with rp49 3′-UTR, Dpt 3′-UTR or when no 3′-UTR was inserted.
Figure 4.
Knocking down Tis11 increased the activity of the reporter construct containing CecA1 3′-UTR. (A) RT–PCR was performed using specific primers to confirm the knockdown efficiency of Tis11 in S2* cells treated with EGFP or Tis11 dsRNA. Results shown are representative of three independent experiments. (B) Fluc activity changes after Tis11 RNAi. S2* cells were co-transfected with dsRNA specific for Tis11 or EGFP and a reporter construct inserted with different 3′-UTR. Luciferase activity was assayed as described. Compared in cells transfected with EGFP dsRNA (mock RNAi), the Fluc activity changes in Tis11 knocked down cells is shown. Values represent the mean ± SD of at least four experiments (*P < 0.05; **P < 0.01 for each construct versus both pAC-Fluc and pAC-Fluc-rp49). (C) S2* cells were co-transfected with dsRNA targeting Tis11 or EGFP, human TTP expression plasmid (pAC-TTP-Flag) or control plasmid (pAC- Flag) and a reporter construct inserted with different 3′-UTR. Luciferase activity was assayed as described. Fluc activity measured in cells co-transfected with the pAC-Fluc control vector, EGFP dsRNA and pAC-Flag control expression plasmid is designated as 1. Values represent the mean ± SD of at least three experiments (*P < 0.05; **P < 0.01 for each construct versus pAC-Flag control plasmid).
Taking advantage of the deletion mutants described in Figure 3A, we next investigated which portion of the CecA1 3′-UTR is indispensable for the destabilization by Tis11. Reducing Tis11 expression through RNAi can increase the Fluc activity ∼50% when fragment II or fragment III containing the ARE of CecA1 3′-UTR is inserted. Conversely, Tis11 RNAi had no significant effect when the constructs were inserted with fragment IV or V of CecA1 3′-UTR (Figure 4B). These results show that Tis11 specifically downregulates CecA1 gene expression through the ARE defined in the proximal region of 3′-UTR for mRNA stability control.
To determine whether mammalian TTP can regulate AMP expression, a TTP expression construct was co-transfected into S2* cells with dsRNA targeting Tis11 such that Tis11 was knocked down by RNAi. The expression of mammalian TTP decreased the Fluc activity significantly in S2* cells transfected with reporter construct inserted with CecA1 3′-UTR and rescued the destabilizing effect on reporter mRNAs bearing AREs from mammalian (TNF-α) and Drosophila sources (CecA1) in S2* cells where Tis11 were knocked down by RNAi (Figure 4C). No significant destabilizing effects of TTP on reporter mRNA bearing Dpt 3′-UTR were observed in S2* cells with or without Tis11 knockdown. It appears that the mammalian and Drosophila TTP orthologs share functional similarity regarding the specific regulation of mRNA with CecA1 3′-UTR but not Dpt 3′-UTR.
Tis11 selectively destabilizes CecA1 mRNA and influences the transient expression kinetics
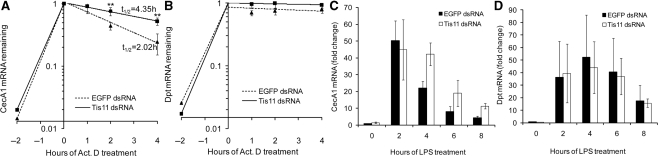
To determine whether Tis11 can differentially reduce the endogenous AMP mRNA stability, Act.D chase studies were performed after S2* cells were treated with dsRNA of Tis11 or EGFP (negative control) for 3 days and stimulated with LPS for 2 h. As shown in Figure 5A, the stability of CecA1 mRNA was significantly increased when Tis11 was knocked down (t1/2 = 2.02 h versus t1/2 = 4.35 h). Conversely, knocking down Tis11 did not change the Dpt mRNA stability (Figure 5B). To further validate the effectiveness of Tis11-regulated mRNA stability on the AMP mRNA expression kinetics, CecA1 and Dpt mRNA expression profiles in S2* cells, in which Tis11 was knocked down by RNAi, were detected after LPS stimulation. As shown in Figure 5C, CecA1 mRNA had a similar fold increase, but remained at a high level longer in Tis11 knocked down cells. The expression profiles of Dpt mRNA were similar whether Tis11 was knocked down or not (Figure 5D).
Figure 5.
The changes in mRNA stability and gene expression profiles of AMPs after Tis11 knockdown. (A and B) S2* cells were transfected with dsRNA specific for Tis11 or EGFP and cultured for 72 h. After exposure of cells to LPS for 2 h, Act.D was added and the preparation incubated for the indicated times. The qRT–PCR was performed to detect CecA1 (A) and Dpt (B) mRNA remaining at each time point, taking the expression level at the time of Act.D addition as 1. (**P<0.01 for EGFP dsRNA versus Tis11 dsRNA at each time point) (C and D) The qRT–PCR analysis of CecA1 (C) or Dpt (D) transcripts in S2* cells after LPS treatment under EGFP or Tis11 RNAi conditions. The fold change of CecA1 or Dpt mRNA was analyzed, taking the expression level in cells transfected with EGFP dsRNA at time point of 0 h as 1. The data presented are mean ± SD of four independent experiments.
These data demonstrate that Tis11 can specifically destabilize CecA1 mRNA and can quickly eliminate CecA1 mRNA to manipulate the transient expression profiles of the potent AMPs when rapidly induced. Meanwhile, Tis11 has no significant effect on mRNA stability and expression profiles of moderately expressed Dpt.
CecA1 mRNA forms an RNA–protein complex with Tis11 in S2* cells
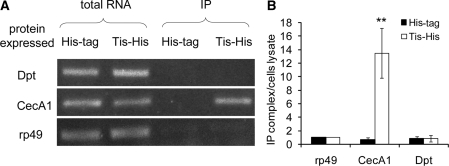
To investigate the activity of Tis11 in specific and direct interaction with AMP mRNA in S2* cells, an IP RT–PCR method was used to measure the amount of mRNA that coprecipitated with Tis11 protein. As shown in Figure 6A, IP with anti-His antibodies dramatically enriched CecA1 mRNA in RNP complexes from cells transfected with the construct expressing the Tis11–His fusion protein, whereas, IP in cells transfected with the construct containing the His tag control had no such effect. In contrast, IP resulted in no enrichment for other mRNAs tested including Dpt (Figure 6A), Def, Dro, AttB, Mtk and Drs in S2* cells expressing blank His tag or Tis11–His fusion protein (data not shown).
Figure 6.
The mRNA analyses of RNP complex IP with Tis11. S2* cells were transfected with construct expressing Tis11-His tag or His tag and incubated for 72 h. Cells were stimulated with LPS for 3 h and then lysed to IP with anti-His tag antibody. (A) RNAs in the cell lysate and the IP complex were identified using RT–PCR with primers specific for CecA1, Dpt or rp49. Results shown are representative of three independent experiments. (B) The CecA1 and Dpt mRNA in the IP complex were analysed by qRT–PCR to evaluate the relative enrichment after co-IP, taking the ratio of rp49 mRNA level in IP samples to cell lysate as 1. Values represent the mean ± SD of three independent experiments (**P < 0.01 for cells expressing Tis-His versus His-tag control).
The changes in CecA1 mRNA after IP were also analyzed using qRT–PCR. After IP with anti-His antibodies in extracts from cells expressing Tis11–His fusion protein, the endogenous CecA1 mRNA was dramatically enriched up to 14-fold as compared with rp49 (Figure 6B). In contrast, none of the other AMP mRNA tested, including Dpt (Figure 6B), Def, Dro, AttB, Mtk and Drs, was enriched (data not shown).
These data demonstrate that Tis11 specifically forms an RNP complex with CecA1 mRNA, but not with Dpt mRNA or any other AMP mRNA examined here in S2* cells. These findings provide strong evidence that the specific interaction between Tis11 and CecA1 mRNA is the essential molecular basis for the differential regulation of genes with distinct expression kinetics.
A crucial role for p38MAPK in differential regulation of AMP mRNA stability mediated by Tis11
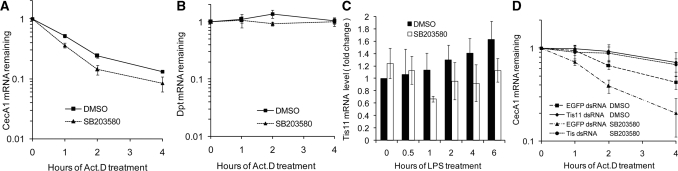
In order to examine the role of p38MAPK in regulating the stability of AMP mRNAs bearing AREs, we investigated the effect of p38MAPK blockade on LPS-induced posttranscriptional regulation of AMP expression. As shown in Figure 7A, inhibition of p38MAPK employing a specific inhibitor, SB203580, decreased the half-life of CecA1 mRNA significantly, whereas Dpt mRNA remained stable and its half-life was unchanged (Figure 7B). These observations demonstrate that LPS-activated p38MAPK stabilizes mRNAs bearing the AREs in S2* cells and that there exist differences in the extent and duration of p38MAPK inhibition between CecA1 and Dpt ARE.
Figure 7.
Effect of p38MAPK inhibition by SB203580 on AMP mRNA stability and Tis11 expression. SB203580 or vehicle (1% DMSO) pretreated S2* cells were incubated with 10 μg/ml LPS for 2 h. Act.D chase studies were performed to detect CecA1 (A) and Dpt (B) mRNA remaining at each time point, taking the expression level at the time of Act.D addition as 1. (C) S2* cells were pretreated for 0.5 h with SB203580 or vehicle. Afterward, cells were incubated with LPS for different times. The fold changes of Tis11 mRNA were detected at each time point, taking the mRNA level of vehicle at 0 h as 1. (D) S2* cells were transfected with dsRNA specific for Tis11 or EGFP and cultured for 72 h. The cells were then treated as described in (A). qRT–PCR was performed to detect CecA1 mRNA remaining at each time point, taking the expression level at the time of Act.D addition as 1.
Through detection of Tis11 expression at mRNA level, it was indicated that LPS stimulation and/or p38MAPK blockade affect the Tis11 expression to some extent but not significantly (Figures 7C). To determine the involvement of Tis11 in p38MAPK-regulated mRNA stabilization, the effects of a p38MAPK blockade on CecA1 mRNA stability were tested in S2* cells with Tis11 knocked down by RNA interference. As shown in Figure 7D, inhibitor SB203580 treatment blocked the stabilization effect of p38MAPK and destabilized CecA1 mRNA to a shorter half-life as compared with the DMSO treated control. However, pretreatment with RNAi knockdown of Tis11 abrogated the destabilization effect of p38MAPK inhibition by SB203580 and increased the half-life of CecA1 mRNA to an extent comparable to the half-life upon Tis11-knockdown-induced stabilization. These results demonstrate that the stabilization effect of LPS-activated p38MAPK on CecA1 mRNA is mediated by the RNA destabilizing protein Tis11 in Drosophila.
DISCUSSION
Drosophila melanogaster can produce a large variety of AMPs, especially when a pathogen entering the body triggers the innate immune system, as shown in previous reports with Northern or micro array analyses using whole flies (10,12,15). Each AMP has a different antimicrobial spectrum and works together with others against microbial infection (1–3). On the other hand, AMPs have great influence on the resident microbiota community which is important for sustaining host health and must be maintained properly (8,9). The overall effects of AMPs on pathogenic and commensal microbes are attributed to each individual AMP with its distinct antimicrobial spectrum. Undoubtedly, a controllable and dynamic antimicrobial spectrum of an integrated variety of AMPs is a complex event and is dependent upon effectively and differentially regulated expression of each AMP gene displaying distinct expression kinetics upon immune response.
In accordance with previous reports (12,18), we observe that, in S2* cells, the AMP genes were transiently expressed in a characteristic pattern in which the mRNA level increased quickly to its peak and declined afterwards upon immune response (Figure 1 and data not shown). Notably, CecA1 mRNA had the highest peak level and the quickest declining rate among all the AMPs tested, including the moderately expressed Dpt, Def, Dro, AttB, Mtk and Drs. The differences in intensity and duration of gene expression indicate that the AMP genes were not regulated through the same mechanism although they shared a similar transient expression pattern. Comparing various properties of AMPs, we noted that the property distinguishing CecA1 from the other AMPs was its broad spectrum of antimicrobial activity. Previous studies have shown that CecA1 has a broad antimicrobial spectrum against Gram-positive bacteria, Gram-negative bacteria and fungi, while the antimicrobial spectra of other AMPs are quite limited. For example, the Dpt only impacts anti Gram-negative bacteria (1–3,6,7). Therefore, modulating CecA1 expression could have significant impact on the overall antimicrobial spectrum produced by all the AMPs. According to this perspective, differential regulation of genes encoding a functionally distinctive AMP, such as CecA1, is crucial for the host to orchestrate a dedicated and well balanced antimicrobial spectrum using a combination of individual AMPs.
Since gene regulation depends upon the interplay among elements that control gene expression at multiple levels including transcription, mRNA stability and translation, the differences in intensity and time of sustained high level between CecA1 and the other AMPs may be attributed to differential regulation at multiple levels. Previous studies have shown that the differences in the κB motif at the promoters of CecA1 and Dpt are not functionally equal, a fact that can partly explain the different expression profiles (16–18). However, neither expression profiles nor elimination of existing mRNA can be exclusively governed by transcriptional control. In fact, here we show that CecA1 mRNA is much more unstable than Dpt as demonstrated using Act.D chase studies for mRNA half-life measurement. These experiments partly explain why CecA1 is eliminated more quickly in LPS-elicited transient expression. In addition, the mRNA decay rate of each AMP was not altered whether or not S2* cells were stimulated with LPS. This was confirmed by additional experiments with reporter constructs containing Dpt or CecA1 3′-UTR, in which the Fluc activity was regulated by the inserted 3′-UTR but not affected by LPS treatment (data not shown). These results indicate that the decay rates of CecA1 and Dpt mRNAs are maintained in a constitutive manner and that the sustained pattern of mRNA stability of both AMPs is under posttranscriptional regulatory control upon LPS stimulation.
The steady decay rate of unstable mRNA results in the effective and constant elimination of mRNA. The expression of low stability genes means quick synthesis and quick mRNA decay and consumes more energy, which is also important for the host. However, compared with induction-dependent regulation, constitutive instability of mRNA is an extremely fast and effective mode for negative control of transient gene expression upon immune response. Of note, proper down-modulation of immunity is critical for protective immunity and health (8,9,42–44), and recent work has shown that hyperactivated or prolonged immune responses, including the expression of AMPs, are detrimental to the host, partly because of an altered commensal microbe population (8,9). A series of negative regulators that control the intensity and duration of AMP expression, mainly at the transcriptional level, have been identified. In extracellular compartments, hemocytes phagocytose microbes and some secreted PGRPs for enzymatic degradation of peptidoglycan (45,46). Inside a host cell, Drosophila Wnt inhibitor of Dorsal (wnt D), Defense repressor 1 (Dnr1), caspar, PIMS and rudra/pirk downregulate the Toll or IMD pathways, the major regulators of the immune response in cytoplasm of Drosophila (43,44,47–50). In the nucleus, AP-1 and STAT complex, activated by the JNK pathway, inhibit AMP expression through removing Relish from the promoter (51). In addition to those transcriptional down-modulations, posttranscriptional events, particularly the instability of specific mRNAs reported here, are also important determinants of the downregulation of AMP gene expression. Given that CecA1 has a more potent response and a broader spectrum of antimicrobial activity than do the other AMPs, differential instability control may play an important role in the strict regulation of its gene expression to avoid excessive effects harmful to resident microbiota and deleterious to host health. Conversely, the moderately expressed Dpt with activity against Gram-negative bacteria may have a less deleterious effect upon the host. Hence, it may not be necessary to eliminate Dpt mRNA quickly with an mRNA instability mechanism. The difference in mRNA stability between AMPs with different antimicrobial spectra provides an effective approach for exact control of AMP gene expression contributing to the integrated antimicrobial activity.
In the 3′-UTR of a variety of transiently expressed immediate response genes there is an ARE involved in the posttranscriptional regulation (20–23,25,26). It is highly conserved throughout evolution and can be found in species ranging from yeast and insects to mammals. Both CecA1 and Dpt contain AU-rich sequences in the 3′-UTR, which likely serve as cis-acting elements and may be involved in posttranscriptional regulation. Our results illustrate that CecA1 3′-UTR, mainly through the ARE in the proximal region, can effectively accelerate reporter mRNA decay and decrease reporter mRNA level as well as reporter Fluc activity. In most cases, the magnitude of the decrease in Fluc activity was directly correlated with the corresponding decrease in Fluc mRNA level, suggesting that reporter gene expression is primarily dependent upon message stability. These results are consistent with the instability of the CecA1 mRNA and indicate that CecA1 3′-UTR contains a functional ARE, a cis element destabilizing mRNA. However, inserting Dpt 3′-UTR downstream of the reporter has no obvious effect on reporter mRNA stability change, but increases the reporter Fluc activity significantly. This implies that a cis element increasing the translation efficiency may exist within the Dpt 3′-UTR or that additional trans factors may be involved in the modulation of translational efficiency, all of which possibilities should be further investigated. The different roles of CecA1 and Dpt 3′-UTR on mRNA stability and protein expression indicate that 3′-UTR plays important roles in versatile and differential regulation of AMP gene expression.
AREs function as posttranscriptional regulatory elements through interactions with specific binding proteins or microRNA (20,21,26,39). Using RNA interference to screen known RNA binding proteins, we identified Drosophila Tis11, a homolog of mammalian TTP, as a trans-factor controlling CecA1 mRNA stability. Characterized by a tandem CCCH zinc-finger (TZF) domain with highly conserved sequences and spacing, TTP can bind to ARE of unstable mRNAs through TZF domains and can induce mRNA deadenylation, promote degradation of the mRNA body by the exosome complex, or assist the RISC–miRNA complex with targeting mRNA for rapid degradation (39,52–54). Drosophila Tis11 also contains two CCCH zinc-finger domains that are necessary for mammalian TTP to interact with AU-rich elements, destabilizing the mRNA of reporter construct with CecA1 3′-UTR mainly through interaction with the ARE in the proximal region of CecA1 3′-UTR. Moreover, we demonstrated, using an IP RT–PCR method, that CecA1 mRNA and Tis11 were co-precipitated in an RNA–protein complex. It is observed that the ARE in the proximal region of CecA1 3′-UTR contains a UUAUUUAUU sequence, characterized previously as a preferred binding site for mammalian TTP, and indicates that Drosophila Tis11 has a similar binding characteristic to that of mammalian TTP. Although Dpt mRNA 3′-UTR contains the AU-rich sequence, UAUUUUAUU, which also has an optimal affinity to mammalian TTP (32,33), it can increase Fluc reporter activity but cannot be pulled down with Drosophila Tis11 protein using co-IP. This is consistent with the fact that knocking down Tis11 or over-expressing TTP has little effect on Dpt mRNA stability and expression profiles. Perhaps Dpt does not have the same recognition and binding property as mammalian ARE, or additional factors involved in influencing the affinity and specificity of the RNA–protein complex forming with Tis11.
Data from the work of Jing et al. (39) have shown that the Drosophila Tis11 can destabilize reporter mRNA inserted with TNF-α 3′-UTR and the involvement of microRNA is indicated. However, reducing the expression of Dicer1, playing key roles in processing small RNAs in miRNA systems, has no effect on expression of reporters inserted with CecA1 3′-UTR (data not shown) indicating that a RISC–miRNA complex does not participate in CecA1 mRNA stability control. While preparing this article, we noticed that recent work reported by Aurélien et al. (55) demonstrated that Tis11 downregulates CecA1 mRNA stability through acceleration of mRNA deadenylation similar to that of TTP in mammals.
AREs can exert mRNA instability effects but can also confer stabilization of mRNA through the p38MAPK pathway. p38MAPK has been shown to regulate both the translation and the stability of inflammatory mRNAs bearing AREs, including TNF-α, COX-2, GMCSF and VEGF (30,31), in mammalian cells. Previous study has shown that p38MAPK regulates levels of AMP transcripts in Drosophila (56). We have demonstrated in this study that p38MAPK plays a crucial role in regulating the stability of AMP mRNAs bearing the AREs in their 3′-UTR upon LPS activation of p38MAPK in S2* cells. The p38MAPK inhibitor SB203580 can specifically and effectively decrease the half-life of CecA1 mRNA. Of note, our results in this report indicate that the decay rate of CecA1 mRNA remains unaffected upon LPS stimulation as compared to the basal level. Thus, it is suggested that p38MAPK contributes its stabilizing role to a regulatory mechanism coordinating stabilizing and destabilizing regulation to ensure a steady level of degradation rate of the transcript upon LPS stimulation.
The important component of the mechanism of p38MAPK-regulated mRNA stability is a protein that forms the link between the p38MAPK pathway and ARE-containing mRNA. In mammals, the TTP has been suggested to be the protein factor but is not firmly recognized because of evidence for and against its involvement in p38MAPK-mediated stabilization (30,57). Using RNAi knockdown of Tis11, the Drosophila homolog of TTP, we observed that the destabilizing effect of p38MAPK blockade by SB203580 was abrogated. This provides evidence that Tis11 is involved in the regulation of AMP mRNA stability by p38MAPK in S2* cells. Further detection of Tis11 expression at mRNA level indicates that LPS stimulation and/or p38MAPK blockade affects Tis11 expression to some extent but not significantly. It remains unclear as to how p38MAPK regulates AMP mRNA stability mediated by Tis11-AREs interaction. In the future, it would be of interest to determine whether protein phosphorylation of Tis11 affects the regulation of mRNA stability and whether p38MAPK regulates the phosphorylation status of Tis11.
ARE is a highly conserved posttranscriptional regulatory element throughout evolution and can be found in species from yeasts and insects to mammals (24–26). Our previous study (38) has shown the existence of ARE in 3′-UTR of AMP mRNA. We further demonstrate in this report that AREs in 3′-UTR of AMP mRNA are functional elements in the posttranscriptional regulation of AMP gene expression, a mechanism that is extremely important in the innate immunity of insects. Likewise, AREs have been known to play critical roles in the posttranscriptional regulation of gene expression in response to immune stimuli in mammals. It was emphasized and explicated in a recent study that ARE-regulated mRNA stability exerts a strong influence on gene expression, in some cases overriding that of transcriptional control elements, and controlling the expression kinetics of genes encoding inflammatory molecules (58). In addition, the trans-acting protein factor mediating ARE destabilization is Drosophila Tis11, the ortholog of mammalian TTP. Although the AREs of AMP mRNA exert no destabilizing effect in mammalian cells (data not shown), our results support a previous report (39) that the Drosophila Tis11 can destabilize reporter mRNA inserted with TNF-α 3′-UTR. Besides, expression of TTP in S2* cells with Tis11 knockdown can rescue the destabilizing effect on reporter mRNAs bearing AREs. This evidence indicates that the ARE destabilizing trans-acting factors are evolutionarily conserved between invertebrate and mammalian immune systems. Furthermore, we reveal that p38MAPK regulates the stability of the AMP mRNA containing ARE in the 3′-UTR as mediated by Tis11, demonstrating a mechanism strikingly similar to that of p38MAPK-regulated mRNA stabilization in mammalian cells through inhibition of TTP-mediated destabilization in most case (31). Obviously, AREs and relevant regulatory mechanisms have become more complex later in evolution, but the main framework is evolutionarily conserved between invertebrates and mammals and reflects many important mechanisms present in ancestral forms. In particular, evolutionary conservation is manifested in posttranscriptional regulation of gene expression in both fly and mammalian immunity.
Taken together, our results demonstrate that AMPs possessing different antimicrobial spectra exhibit significant differences in gene expression profiles evidently attributed to differential regulation at the posttranscriptional level. While Dpt 3′-UTR has no destabilizing effect, CecA1 3′-UTR is both necessary and sufficient to confer instability through the ARE–Tis11 interaction within the RNP complex. It is further demonstrated that the destabilization is counteracted by p38MAPK which plays a crucial role in stabilizing ARE-bearing mRNAs by inhibiting Tis11-mediated degradation, a posttranscriptional mechanism that is evolutionarily conserved in both fly and mammalian immunity.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China (No. 30570390, No. 30470381); the Shanghai Commission of Science and Technology; Knowledge Innovation Program of the Chinese Academy of Sciences. Funding for open access charge: Knowledge Innovation Program of the Chinese Academy of Sciences.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Professor Ge Baoxue and Professor Jing Qing for the kind provision of Drosophila S2* cells and plasmids of pAc5.1-Flag-V5-His C and pCDNA3.0-myc-TTP. We thank Dr Sheri M. Skinner (University of Texas School of Medicine, TX, USA) for help with preparation of the article.
REFERENCES
- 1.Brennan CA, Anderson KV. Drosophila: the genetics of innate immune recognition and response. Annu. Rev. Immunol. 2004;22:457–483. doi: 10.1146/annurev.immunol.22.012703.104626. [DOI] [PubMed] [Google Scholar]
- 2.Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 3.Uvell H, Engstrom Y. A multilayered defense against infection: combinatorial control of insect immune genes. Trends Genet. 2007;23:342–349. doi: 10.1016/j.tig.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Dushay MS, Eldon ED. Drosophila immune responses as models for human immunity. Am. J. Hum. Genet. 1998;62:10–14. doi: 10.1086/301694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann JA. The immune response of Drosophila. Nature. 2003;426:33–38. doi: 10.1038/nature02021. [DOI] [PubMed] [Google Scholar]
- 6.Samakovlis C, Kimbrell DA, Kylsten P, Engstrom A, Hultmark D. The immune response in Drosophila: pattern of cecropin expression and biological activity. EMBO J. 1990;9:2969–2976. doi: 10.1002/j.1460-2075.1990.tb07489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ekengren S, Hultmark D. Drosophila cecropin as an antifungal agent. Insect Biochem. Mol. Biol. 1999;29:965–972. doi: 10.1016/s0965-1748(99)00071-5. [DOI] [PubMed] [Google Scholar]
- 8.Ryu JH, Kim SH, Lee HY, Bai JY, Nam YD, Bae JW, Lee DG, Shin SC, Ha EM, Lee WJ. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science. 2008;319:777–782. doi: 10.1126/science.1149357. [DOI] [PubMed] [Google Scholar]
- 9.Silverman N, Paquette N. Immunology The right resident bugs. Science. 2008;319:734–735. doi: 10.1126/science.1154209. [DOI] [PubMed] [Google Scholar]
- 10.Lemaitre B, Reichhart JM, Hoffmann JA. Drosophila host defense: differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc. Natl Acad. Sci. USA. 1997;94:14614–14619. doi: 10.1073/pnas.94.26.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tzou P, Reichhart JM, Lemaitre B. Constitutive expression of a single antimicrobial peptide can restore wild-type resistance to infection in immunodeficient Drosophila mutants. Proc. Natl Acad. Sci. USA. 2002;99:2152–2157. doi: 10.1073/pnas.042411999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boutros M, Agaisse H, Perrimon N. Sequential activation of signaling pathways during innate immune responses in Drosophila. Dev. Cell. 2002;3:711–722. doi: 10.1016/s1534-5807(02)00325-8. [DOI] [PubMed] [Google Scholar]
- 13.Orphanides G, Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–451. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- 14.Engstrom Y. Induction and regulation of antimicrobial peptides in Drosophila. Dev. Comp. Immunol. 1999;23:345–358. doi: 10.1016/s0145-305x(99)00016-6. [DOI] [PubMed] [Google Scholar]
- 15.Hedengren-Olcott M, Olcott MC, Mooney DT, Ekengren S, Geller BL, Taylor BJ. Differential activation of the NF-kappaB-like factors Relish and Dif in Drosophila melanogaster by fungi and Gram-positive bacteria. J. Biol. Chem. 2004;279:21121–21127. doi: 10.1074/jbc.M313856200. [DOI] [PubMed] [Google Scholar]
- 16.Busse MS, Arnold CP, Towb P, Katrivesis J, Wasserman SA. A kappaB sequence code for pathway-specific innate immune responses. EMBO J. 2007;26:3826–3835. doi: 10.1038/sj.emboj.7601798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gross I, Georgel P, Kappler C, Reichhart JM, Hoffmann JA. Drosophila immunity: a comparative analysis of the Rel proteins dorsal and Dif in the induction of the genes encoding diptericin and cecropin. Nucleic Acids Res. 1996;24:1238–1245. doi: 10.1093/nar/24.7.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han ZS, Ip YT. Interaction and specificity of Rel-related proteins in regulating Drosophila immunity gene expression. J. Biol. Chem. 1999;274:21355–21361. doi: 10.1074/jbc.274.30.21355. [DOI] [PubMed] [Google Scholar]
- 19.Tanji T, Hu X, Weber AN, Ip YT. Toll and IMD pathways synergistically activate an innate immune response in Drosophila melanogaster. Mol. Cell Biol. 2007;27:4578–4588. doi: 10.1128/MCB.01814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bevilacqua A, Ceriani MC, Capaccioli S, Nicolin A. Post-transcriptional regulation of gene expression by degradation of messenger RNAs. J. Cell Physiol. 2003;195:356–372. doi: 10.1002/jcp.10272. [DOI] [PubMed] [Google Scholar]
- 21.Guhaniyogi J, Brewer G. Regulation of mRNA stability in mammalian cells. Gene. 2001;265:11–23. doi: 10.1016/s0378-1119(01)00350-x. [DOI] [PubMed] [Google Scholar]
- 22.Cheadle C, Fan J, Cho-Chung YS, Werner T, Ray J, Do L, Gorospe M, Becker KG. Stability regulation of mRNA and the control of gene expression. Ann. NY Acad. Sci. 2005;1058:196–204. doi: 10.1196/annals.1359.026. [DOI] [PubMed] [Google Scholar]
- 23.Khabar KS. The AU-rich transcriptome: more than interferons and cytokines, and its role in disease. J. Interferon Cytokine Res. 2005;25:1–10. doi: 10.1089/jir.2005.25.1. [DOI] [PubMed] [Google Scholar]
- 24.Vasudevan S, Peltz SW. Regulated ARE-mediated mRNA decay in Saccharomyces cerevisiae. Mol. Cell. 2001;7:1191–1200. doi: 10.1016/s1097-2765(01)00279-9. [DOI] [PubMed] [Google Scholar]
- 25.Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 26.Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 28.Ma WJ, Cheng S, Campbell C, Wright A, Furneaux H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem. 1996;271:8144–8151. doi: 10.1074/jbc.271.14.8144. [DOI] [PubMed] [Google Scholar]
- 29.Abdelmohsen K, Lal A, Kim HH, Gorospe M. Posttranscriptional orchestration of an anti-apoptotic program by HuR. Cell Cycle. 2007;6:1288–1292. doi: 10.4161/cc.6.11.4299. [DOI] [PubMed] [Google Scholar]
- 30.Frevel MA, Bakheet T, Silva AM, Hissong JG, Khabar KS, Williams BR. p38 Mitogen-activated protein kinase-dependent and -independent signaling of mRNA stability of AU-rich element-containing transcripts. Mol. Cell Biol. 2003;23:425–436. doi: 10.1128/MCB.23.2.425-436.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dean JL, Sully G, Clark AR, Saklatvala J. The involvement of AU-rich element-binding proteins in p38 mitogen-activated protein kinase pathway-mediated mRNA stabilisation. Cell Signal. 2004;16:1113–1121. doi: 10.1016/j.cellsig.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 32.Worthington MT, Pelo JW, Sachedina MA, Applegate JL, Arseneau KO, Pizarro TT. RNA binding properties of the AU-rich element-binding recombinant Nup475/TIS11/tristetraprolin protein. J. Biol. Chem. 2002;277:48558–48564. doi: 10.1074/jbc.M206505200. [DOI] [PubMed] [Google Scholar]
- 33.Brewer BY, Malicka J, Blackshear PJ, Wilson GM. RNA sequence elements required for high affinity binding by the zinc finger domain of tristetraprolin: conformational changes coupled to the bipartite nature of Au-rich MRNA-destabilizing motifs. J. Biol. Chem. 2004;279:27870–27877. doi: 10.1074/jbc.M402551200. [DOI] [PubMed] [Google Scholar]
- 34.Zhuang ZH, Zhou Y, Yu MC, Silverman N, Ge BX. Regulation of Drosophila p38 activation by specific MAP2 kinase and MAP3 kinase in response to different stimuli. Cell Signal. 2006;18:441–448. doi: 10.1016/j.cellsig.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 35.Leulier F, Parquet C, Pili-Floury S, Ryu JH, Caroff M, Lee WJ, Mengin-Lecreulx D, Lemaitre B. The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat. Immunol. 2003;4:478–484. doi: 10.1038/ni922. [DOI] [PubMed] [Google Scholar]
- 36.Kaneko T, Goldman WE, Mellroth P, Steiner H, Fukase K, Kusumoto S, Harley W, Fox A, Golenbock D, Silverman N. Monomeric and polymeric gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity. 2004;20:637–649. doi: 10.1016/s1074-7613(04)00104-9. [DOI] [PubMed] [Google Scholar]
- 37.Petersen RB, Lindquist S. Regulation of HSP70 synthesis by messenger RNA degradation. Cell Regul. 1989;1:135–149. doi: 10.1091/mbc.1.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen HX, Li Y, Jiang ZZ, Qu XM, Yang SL, Ma WJ. The existence of a putative post-transcriptional regulatory element in 3'-UTR of Drosophila antibacterial peptide diptericin's; mRNA. FEBS Lett. 2004;561:181–185. doi: 10.1016/S0014-5793(04)00161-9. [DOI] [PubMed] [Google Scholar]
- 39.Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, Di Padova F, Lin SC, Gram H, Han J. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 40.Borgeson CD, Samson ML. Shared RNA-binding sites for interacting members of the Drosophila ELAV family of neuronal proteins. Nucleic Acids Res. 2005;33:6372–6383. doi: 10.1093/nar/gki942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park SJ, Yang ES, Kim-Ha J, Kim YJ. Down regulation of extramacrochaetae mRNA by a Drosophila neural RNA binding protein Rbp9 which is homologous to human Hu proteins. Nucleic Acids Res. 1998;26:2989–2994. doi: 10.1093/nar/26.12.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schneider DS. How and why does a fly turn its immune system off? PLoS Biol. 2007;5:e247. doi: 10.1371/journal.pbio.0050247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lhocine N, Ribeiro PS, Buchon N, Wepf A, Wilson R, Tenev T, Lemaitre B, Gstaiger M, Meier P, Leulier F. PIMS modulates immune tolerance by negatively regulating Drosophila innate immune signaling. Cell Host Microbe. 2008;4:147–158. doi: 10.1016/j.chom.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 44.Gordon MD, Dionne MS, Schneider DS, Nusse R. WntD is a feedback inhibitor of Dorsal/NF-kappaB in Drosophila development and immunity. Nature. 2005;437:746–749. doi: 10.1038/nature04073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bischoff V, Vignal C, Duvic B, Boneca IG, Hoffmann JA, Royet J. Downregulation of the Drosophila immune response by peptidoglycan-recognition proteins SC1 and SC2. PLoS Pathog. 2006;2:e14. doi: 10.1371/journal.ppat.0020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zaidman-Remy A, Herve M, Poidevin M, Pili-Floury S, Kim MS, Blanot D, Oh BH, Ueda R, Mengin-Lecreulx D, Lemaitre B. The Drosophila amidase PGRP-LB modulates the immune response to bacterial infection. Immunity. 2006;24:463–473. doi: 10.1016/j.immuni.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 47.Aggarwal K, Rus F, Vriesema-Magnuson C, Erturk-Hasdemir D, Paquette N, Silverman N. Rudra interrupts receptor signaling complexes to negatively regulate the IMD pathway. PLoS Pathog. 2008;4:e1000120. doi: 10.1371/journal.ppat.1000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guntermann S, Primrose DA, Foley E. Dnr1-dependent regulation of the Drosophila immune deficiency signaling pathway. Dev. Comp. Immunol. 2009;33:127–134. doi: 10.1016/j.dci.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 49.Kleino A, Myllymaki H, Kallio J, Vanha-aho LM, Oksanen K, Ulvila J, Hultmark D, Valanne S, Ramet M. Pirk is a negative regulator of the Drosophila Imd pathway. J. Immunol. 2008;180:5413–5422. doi: 10.4049/jimmunol.180.8.5413. [DOI] [PubMed] [Google Scholar]
- 50.Kim M, Lee JH, Lee SY, Kim E, Chung J. Caspar, a suppressor of antibacterial immunity in Drosophila. Proc. Natl Acad. Sci. USA. 2006;103:16358–16363. doi: 10.1073/pnas.0603238103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim LK, Choi UY, Cho HS, Lee JS, Lee WB, Kim J, Jeong K, Shim J, Kim-Ha J, Kim YJ. Down-regulation of NF-kappaB target genes by the AP-1 and STAT complex during the innate immune response in Drosophila. PLoS Biol. 2007;5:e238. doi: 10.1371/journal.pbio.0050238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 53.Wilusz CJ, Wormington M, Peltz SW. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 54.Lai WS, Carballo E, Strum JR, Kennington EA, Phillips RS, Blackshear PJ. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol. Cell Biol. 1999;19:4311–4323. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lauwers A, Twyffels L, Soin R, Wauquier C, Kruys V, Gueydan C. Post-transcriptional Regulation of Genes Encoding Anti-microbial Peptides in Drosophila. J. Biol. Chem. 2009;284:8973–8983. doi: 10.1074/jbc.M806778200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Han ZS, Enslen H, Hu X, Meng X, Wu IH, Barrett T, Davis RJ, Ip YT. A conserved p38 mitogen-activated protein kinase pathway regulates Drosophila immunity gene expression. Mol. Cell Biol. 1998;18:3527–3539. doi: 10.1128/mcb.18.6.3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sandler H, Stoecklin G. Control of mRNA decay by phosphorylation of tristetraprolin. Biochem. Soc. Trans. 2008;36:491–496. doi: 10.1042/BST0360491. [DOI] [PubMed] [Google Scholar]
- 58.Hao S, Baltimore D. The stability of mRNA influences the temporal order of the induction of genes encoding inflammatory molecules. Nat. Immunol. 2009;10:281–288. doi: 10.1038/ni.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.