Abstract
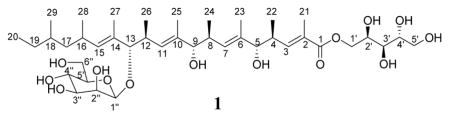
A new polyketide glycoside, bionectriol A (1), was produced by a fungal culture of Bionectria sp., which was isolated from a fungus garden of the fungus-growing ant Apterostigma dentigerum, in Costa Rica. The structure of bionectriol A was determined mainly through NMR and mass spectroscopic data, as well as UV and IR spectra. The relative configurations of the main chain, the pyranohexose, and the pentitol moiety were elucidated by 1H-1H coupling constants and ROESY NMR spectral analysis.
Keywords: polyketide, glycoside, fungus-growing ant
Microbes that mediate complex ecological interactions among “higher” organisms have recently emerged as rich sources of novel bioactive metabolites.1,2,3 In one closely studied system, fungus-growing ants (Attini: Formicidae) cultivate a food fungus in specialized gardens (Agaricales: Basidiomycota: Lepiotaceae or Pterulaceae). These ants were recently shown to harbor mutualistic bacteria in the genus Pseudonocardia (Actinobacteria), which produce antibiotics that target Escovopsis sp. (Ascomycota), a specialized fungal pathogen of the ants’ fungal gardens.4,5 The ant-fungus mutualism originated approximately 50 million years ago in the Amazon Basin and evolved into more than 230 species of ants, each associated with lineages of mutualistic fungi, antagonistic fungi (Escovopsis), and mutualist bacteria (Pseudonocardia).5,6 The diversity of the known microbial symbionts associated with fungus-growing ants, coupled with evidence that they produce diverse secondary metabolites (especially Pseudonocardia),5,7 makes this ancient mutualism a potentially rich but unexplored source of natural products.
Even though the fungus-growing ant system has been studied for more than 130 years,8 secondary metabolites from the microorganisms in this niche have received only limited attention.9 However, our recent study on one of the fungus-growing ant symbiotic systems, Apterostigma dentigerum in Panama, revealed an antifungal compound produced by the symbiotic actinomycete, Pseudonocardia sp. This new antifungal agent, a cyclic depsipeptide bearing unusual amino acids, selectively inhibits the antagonistic fungus over the mutualistic fungus, indicating that these symbioses are promising sources for bioactive natural products.10
Apterostigma occur throughout Central and South American and often nest in, or under decaying logs or in root banks near streams. Unlike most Attine ants that cultivate litter-decomposing fungi (Basidiomycota: Agaricales), Apterostigma grow a distantly related wood-gardening Basidiomycete (Pterulaceae).11,12 The ants use insect feces and dead woody material to cultivate their food fungal crop.13 In addition to the described fungal and actinomycete symbionts, it is possible that other microorganisms as yet unknown from the system could serve as additional sources for compound discovery. Several studies have shown that gardens from different fungus-growing ant species host a variety of microorganisms with unknown roles.14,15,16 Here we examine a fungus with an unknown role that was isolated from the garden of the fungus-growing ant Apterostigma dentigerum.
We have confirmed earlier findings14,15,16 that fungus-growing ant gardens harbor additional fungal strains besides the cultivar and parasitic fungi. We isolated a strain, CC061026-06, from a nest of the attine ant Apterostigma dentigerum in Costa Rica. 28S rRNA sequencing and phylogenetic analysis were conducted to identify this strain. High bootstrap support indicates that its closest relative is Bionectria ochroleuca. Additionally, phylogenetic analysis shows that the strain is closely related to other strains of Nectria/Bionectria, and that this group is sister to Hydropisphaera. Bionectria is neither a known crop cultivated by A. dentigerum nor a known fungal pathogen; the exact role of CC061026-06 within the ant-fungal mutualism is unknown. Species of Bionectria have, however, been identified from other insect systems including several wood-feeding bark beetle species17,18 and some strains were isolated from oral secretions.18 As part of our effort to probe the metabolic repertoire of the microorganisms in fungus-growing ant systems, we investigated the secondary metabolite profile of this fungal strain, Bionectria sp. Here we report bionectriol A, a previously unreported polyketide glycoside, produced by strain CC061026-06.
The fungal strain was cultured in potato dextrose broth, with agitation at 250 rpm, at 30 °C for 12 days. An ethyl acetate extract of cultured cells and supernatant (total volume 600 mL) was subjected to reversed-phase flash column chromatography and final purification by HPLC to yield 3.7 mg bionectriol A (1). Bionectriol A was obtained as a viscous oil with the molecular formula C40H70O14, as confirmed by ESIHRMS (obsd [M+Na]+ at m/z 797.4670, calcd [M+Na]+ 797.4663). This molecular formula was also supported by 1H and 13C NMR data (Table 1).
Table 1.
NMR Data for bionectriol A (1) in CD3OD.
| C/H | δHa | mult (J in Hz) | δCb | #H |
|---|---|---|---|---|
| 1 | 168.5 | C | ||
| 2 | 127.5 | C | ||
| 3 | 6.78 | dd (10.0, 1.5) | 146.5 | CH |
| 4 | 2.73 | m | 37.0 | CH |
| 5 | 3.82 | m | 82.3 | CH |
| 6 | 136.0 | C | ||
| 7 | 5.32 | d (9.5) | 133.0 | CH |
| 8 | 2.63 | m | 35.7 | CH |
| 9 | 3.68 | d (9.5) | 83.0 | CH |
| 10 | 134.7 | C | ||
| 11 | 5.37 | d (9.0) | 133.5 | CH |
| 12 | 2.72 | m | 34.2 | CH |
| 13 | 3.95 | d (9.5) | 87.3 | CH |
| 14 | 139.6 | C | ||
| 15 | 5.25 | d (9.5) | 129.9 | CH |
| 16 | 2.60 | m | 30.0 | CH |
| 17 | 1.28 | m | 44.5 | CH2 |
| 18 | 1.30 | m | 33.0 | CH |
| 19 | 1.15 | m | 26.0 | CH2 |
| 20 | 0.86 | t (6.5) | 10.8 | CH3 |
| 21 | 1.88 | d (1.5) | 11.8 | CH3 |
| 22 | 0.85 | m | 15.4 | CH3 |
| 23 | 1.67 | s | 10.0 | CH3 |
| 24 | 0.77 | d (7.0) | 16.6 | CH3 |
| 25 | 1.66 | s | 10.0 | CH3 |
| 26 | 0.80 | d (7.0) | 16.2 | CH3 |
| 27 | 1.60 | d (1.0) | 10.0 | CH3 |
| 28 | 0.98 | d (6.5) | 21.3 | CH3 |
| 29 | 0.84 | m | 18.4 | CH3 |
| 1′a | 4.18 | dd (11.0, 5.5) | 66.6 | CH2 |
| 1′b | 4.26 | dd (11.0, 7.5) | ||
| 2′ | 4.15 | ddd (7.5, 5.5, 1.5) | 68.0 | CH |
| 3′ | 3.53 | m | 71.2 | CH |
| 4′ | 3.71 | ddd (7.5, 6.0, 3.5) | 71.7 | CH |
| 5′a | 3.81 | dd (11.0, 3.5) | 63.9 | CH2 |
| 5′b | 3.64 | dd (11.0, 6.0) | ||
| 1″ | 4.35 | bs | 96.3 | CH |
| 2″ | 3.74 | m | 73.8 | CH |
| 3″ | 3.35 | dd (9.5, 3.0) | 74.5 | CH |
| 4″ | 3.53 | m | 67.6 | CH |
| 5″ | 3.04 | ddd (9.5, 5.5, 2.5) | 77.0 | CH |
| 6″a | 3.72 | m | 62.0 | CH2 |
| 6″b | 3.86 | dd (12.0, 2.5) |
600 MHz.
150 MHz.
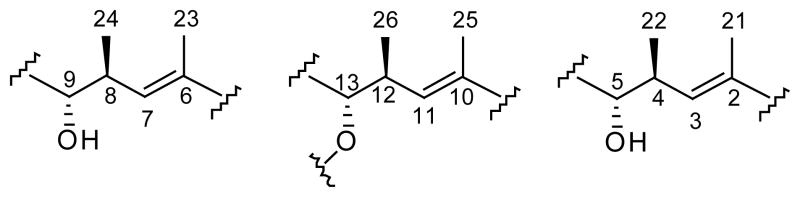
The planar structure of bionectriol A was elucidated mainly by two-dimensional NMR experiments, including 1H-1H COSY, TOCSY, HSQC, and HMBC spectra. The 13C NMR and the HSQC spectra suggested the presence of one carbonyl carbon, four methine and four quaternary sp2 resonances, eleven oxygenated methines and three oxygenated methylenes, two aliphatic methylene carbons, five aliphatic methine carbons, and 10 methyl groups. A network of COSY correlations between H-9 and H-8, between H-8 and H3-24, between H-8 and H-7, and between H-7 and H3-25 (long-range COSY correlation) constructed the C6-substructure shown in Figure 1. In a similar manner, two more repeating C6-substructures were assigned.
Figure 1.
The repeating C6 substructures in the structure elucidation of bionectriol A (1).
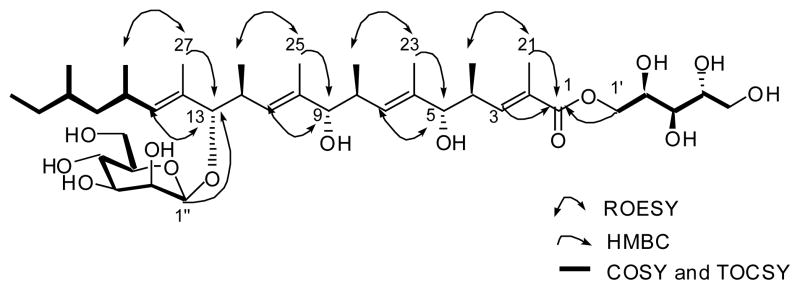
The HMBC correlations from H3-23 to C-5, from H3-25 to C-9, and from H3-27 to C-13 connected these three consecutive and repeating units (Figure 2). The termini of the repeating chain were assigned by the consecutive COSY and TOCSY correlations through H-15 to H-20 and the HMBC correlations from H-3 and H3-21 to C-1 (Figure 2).
Figure 2.
Selected 2D NMR correlations for bionectriol A (1).
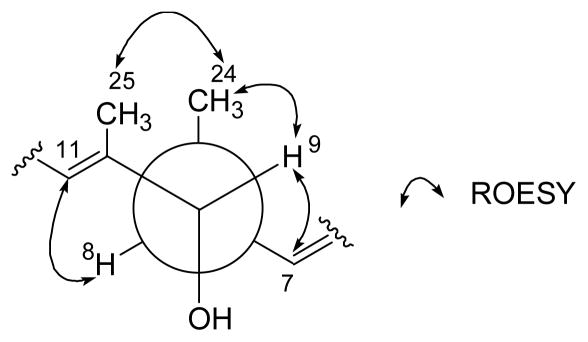
The pentitol, the five-carbon polyhydroxy ester fragment, was identified on the basis of the COSY, TOCSY, and HMBC spectral analysis. This pentitol was connected to the C-1 ester carbonyl carbon (Figure 2). The hexopyranose moiety was also elucidated by the COSY and HMBC correlations. Attachment of the sugar at C-13 was deduced from an HMBC correlation from the acetal proton (H-1″, δH 4.35, δC 96.2). This sugar was identified as mannose by the 1H-1H coupling constant and ROESY spectral interpretation. The coupling constant between H-4″ and H-5″ (J = 9.0 Hz) established their axial-axial relationship. The ROESY correlations from H-1″ to H-3″ and H-5″ indicated that these protons were axial. An additional ROESY correlation between H-1″ and H-2″ showed that H-2″ is equatorial. The axial configuration of the anomeric proton assigned the glycosidic linkage as β-configuration, which was further supported by the one-bond C-H coupling constant of 157 Hz between the anomeric carbon and proton.19 The geometry of all the double bonds in 1 was assigned as E based on the ROESY correlations as shown in Figure 2. The relative configuration between the hydroxy group at C-9 and the methyl group (C-24) at C-8 was established by 1H-1H coupling constant and ROESY correlations at these stereocenters. The large coupling constant (9.5 Hz) between H-8 and H-9 defined their anti-relationship. Since H-8 and H-9 were fixed as anti, the ROESY couplings shown in Figure 3 also defined an anti relationship between the methyl group (C-24) and the hydroxy group at C-9. By a similar logic, the other two repeating units also have an anti relationship between their aliphatic methyl groups (C-22 and C-26) and the hydroxy group and the ester group, respectively.
Figure 3.
Rotamer of C-8 and C-9 stereogenic centers with ROESY correlations.
To determine the relative configuration of the pentitol moiety, we acquired 1H-1H coupling constants by utilizing selective 1D TOCSY spectra. The coupling constants and the carbon chemical shifts of the three consecutive stereogenic centers (2″, 3″, and 4″) indicated that the pentitol moiety has 2″R*, 3″R*, and 4″R* relative configuration based on the published values of the identical pentitol moiety in roselipin 1B.20
The structure of bionectriol A suggests that it is most likely produced by a polyketide synthase (PKS) from one molecule of acetyl-coenzyme A (acetyl-CoA) and nine molecules of malonyl-CoA. The malonyl CoAs are presumably methylated by C-methylation (CMeT) domains utilizing an S-adenosyl methionine (SAM) cofactor after each ketosynthase step, as has been documented in other fungal PKS systems.21 The structure of the core of bionectriol A is consistent with an iterative PKS assembly model, in which a single pair of modules is responsible for incorporation of the third and fourth, fifth and sixth, and seventh and eighth two-carbon building blocks. We speculate that, after the bionectriol A core has been generated by the iterative PKS machinery, it undergoes mannosylation on the C-13 hydroxyl and esterification to a pentitol to yield 1.
The closest known structural relatives of bionectriol A (1) are roselipins 1A and 1B, which were previously isolated from a marine fungal isolate of Gliocladium roseum KF-1040, the anamorph of B. ochroleuca.20,22,23 The roselipins, which are diastereomers differing in configuration at C2′ and C4′, differ from bionectriol A by a double bond between C-14 and C-15. Bionectriol A is mannosylated, as are the roselipins.
The biological activity of 1 is currently under evaluation. Co-plating strain CC061026-06 with a variety of fungal isolates (Escovopsis spp.) revealed no significant antifungal activity. Also, an antifungal assay with pure bionectriol A against Candida albicans ATCC # 10231 showed no significant inhibition.
Bionectriol A represents a class of glycosylated, polyunsatured polyols whose biological and ecological functions are unknown. While the other members of this class, the roselipins, were found in Gliocladium isolates associated with seaweed, bionectriol A was derived from an different context, a fungus-growing ant colony. These findings suggest that this class of compounds may play a role in interspecies interactions, although their activity remains unclear. Further investigation of the community of “bystander” microbes associated with fungus-growing ants as well as the small molecules they produce, will be needed to understand the chemical ecology of this symbiosis.
Experimental section
General experimental procedures
Optical rotation was measured on a Jasco P-2000 polarimeter with a 5 cm cell. A UV-visible spectrum was acquired in methanol on an Amersham Ultrospec 5300 Pro UV/vis spectrophotometer with a path length of 1 cm. An IR spectrum was obtained on a Perkin Elmer 1600 FT-IR instrument. All NMR spectra were collected on a Varian Oxford AS600 instrument, operating at 600 MHz. Reversed-phase liquid chromatography-mass spectroscopy was carried out on an Agilent 1200 series LC instrument coupled to an Agilent 6130 Quadrupole mass spectrometer. High-resolution mass spectra were obtained on an Ultima Q-TOF instrument at the Mass Spectrometry Center, University of Illinois. HPLC was carried out using an Agilent 1100 series apparatus.
Collection and identification of fungal isolate
Filamentous fungi were isolated from a colony of Apterostigma dentigerum ants collected at the La Selva Biological Station in Costa Rica (10° 25′ 53″ N, 84° 00′ 13″ W) in October 2006. The isolation was conducted by plating garden substrate on potato dextrose agar plates containing antibacterial antibiotics, following previously published methods.4 Fungal genomic DNA was isolated by use of the Epicentre MasterPure DNA purification kit protocol for plant tissue, without modification. A 900-basepair fragment of the gene encoding the 28S ribosomal RNA was amplified by polymerase chain reaction using primers LR5 (5′-TCC TGA GGG AAA CTT CG-3′) and LR0R (5′-ACC CGC TGA ACT TAA GC-3′), as described.24 The PCR product was sequenced at the Dana Farber/Harvard Cancer Center DNA Sequencing Facility using the same primers. The phylogenetic placement for strain CC061026-06 was assessed using maximum likelihood (ML) analysis of ~548 base pairs from 28S rDNA sequences. For this analysis we used 22 representative members of Hypocreomycetidae (Ascomycota), 19 of which were from the order Hypocreales.25 Samples were first aligned in ClustalW using default parameters,26 and ML analysis was performed using GARLI (Genetic Algorithm for Rapid Likelihood Inference).27 Default settings were used to estimate the substitution model (general time reversible model), base pair frequencies, and among-site rate variation (gamma distribution).28 Characters were all treated as unordered, and gaps as missing data. Bootstrap support for ML was calculated for internal branches after 100 pseudoreplicates. Output files from GARLI were imported into TREEVIEW to order taxa, display bootstrap values, and root the tree.23
Extraction, isolation, and characterization
The fungus was cultured in potato dextrose broth, with agitation, for 12 days at 30 °C. Fungal tissue and culture supernatant (total volume of 600 mL) were extracted with ethyl acetate (2 L). The extract was dried with sodium sulfate and filtered, and the solvent was removed in vacuo. The crude extract was subjected to reversed-phase flash column chromatography on C18 resin using a stepped gradient from 20% MeOH in H2O to 100% MeOH. Fractions from this column containing bionectriol A (1) on the basis of LC-MS analysis were further purified by HPLC using an Altima C18 semipreparative column at a flow rate of 5 mL/min under isocratic conditions (35% acetonitrile in H2O) to yield 3.7 mg bionectriol A.
Bionectriol A (1): amorphous solid; [α]D +19 (c 0.31; MeOH); UV (MeOH) λmax (log ε) 216 (sh,4.0) nm; IR (NaCl disk) νmax 3330, 2963, 1702, 1457, 1080, 1029 cm−1; 1H NMR data, see Table 1; 13C NMR data, see Table 1; HRMS m/z found 797.4670 [M + Na]+ (calcd for C40H70O14Na, 797.4663).
Supplementary Material
Acknowledgments
This research was supported by N.I.H. grant CA24487 to J.C. and NSF CAREER grant 747002 to C.R.C. E.F. is a fellow of the Fannie and John Hertz Foundation.
Footnotes
Supplementary Data. Experimental methods, 1H, 13C, 2D NMR spectra of 1, and phylogenetic analysis of the strain CC061026-06 are available.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Handelsman J. Microbiol Mol Biol Rev. 2004;68:669–685. doi: 10.1128/MMBR.68.4.669-685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scott JJ, Oh DC, Yuceer MC, Klepzig KD, Clardy J, Currie CR. Science. 2008;322:63. doi: 10.1126/science.1160423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oh DC, Scott JJ, Currie CR, Clardy J. Org Lett. 2009;11:633–636. doi: 10.1021/ol802709x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Currie CR, Scott JA, Summerbell RC, Malloch D. Nature. 1999;398:701–704. [Google Scholar]
- 5.Currie CR, Poulsen M, Mendenhall J, Boomsma JJ, Billen J. Science. 2006;311:81–83. doi: 10.1126/science.1119744. [DOI] [PubMed] [Google Scholar]
- 6.Schultz TR, Brady SG. Proc Natl Acad Sci USA. 2008;105:5435–5440. doi: 10.1073/pnas.0711024105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poulsen M, Erhardt DP, Molinaro DJ, Lin TL, Currie CR. PLoS One. 2007;9:e960. doi: 10.1371/journal.pone.0000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belt T. The Naturalist in Nicaragua. E. Bumpas; London: 1874. p. 403. [Google Scholar]
- 9.Wang Y, Mueller UG, Clardy J. J Chem Ecol. 1999;25:935–941. [Google Scholar]
- 10.Oh D-C, Poulsen M, Currie CR, Clardy J. Nat Chem Biol. 2009;5:391–393. doi: 10.1038/nchembio.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chapela IH, Rehner SA, Schultz TR, Mueller UG. Science. 1994;266:1691–1694. doi: 10.1126/science.266.5191.1691. [DOI] [PubMed] [Google Scholar]
- 12.Villesen P, Mueller UG, Schultz TR, Adams RM, Bouck AC. Evolution. 2004;58:2252–2265. doi: 10.1111/j.0014-3820.2004.tb01601.x. [DOI] [PubMed] [Google Scholar]
- 13.Hölldobler B, Wilson EO. The Ants. Belknap Press; Boston: 1990. [Google Scholar]
- 14.Currie CR, Mueller UG, Malloch D. Proc Natl Acad Sci USA. 1999;96:7998–8002. doi: 10.1073/pnas.96.14.7998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodrigues A, Bacci M, Jr, Mueller UG, Ortiz A, Pagnocca FC. Microb Ecol. 2008;56:604–614. doi: 10.1007/s00248-008-9380-0. [DOI] [PubMed] [Google Scholar]
- 16.Fisher PJ, Stradling DJ, Sutton BC, Petrini LE. Mycol Res. 1996;100:541–546. [Google Scholar]
- 17.Linnakoski R, de Beer ZW, Rousi M, Niemelä P, Pappinen A, Wingfield MJ. Mycol Res. 2008;112:1475–1488. doi: 10.1016/j.mycres.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 18.Cardoza YJ, Vasanthakumar A, Suazo A, Raff KF. Entomol Exp Appl. 2009;131:138–147. [Google Scholar]
- 19.Pretsch E, Bühlmann P, Affolter C. Structure determination of organic compounds-tables of spectraol data. Springer; New York: 2000. p. 163. [Google Scholar]
- 20.Tabata N, Ohyama Y, Tomoda H, Abe T, Namikoshi M, Omura S. J Antibiot. 1999;52:815–826. doi: 10.7164/antibiotics.52.815. [DOI] [PubMed] [Google Scholar]
- 21.Cox RJ. Org Biomol Chem. 2007;5:2010–2026. doi: 10.1039/b704420h. [DOI] [PubMed] [Google Scholar]
- 22.Tomoda H, Ohyama Y, Abe T, Tabata N, Namikoshi M, Yamaguchi Y, Masuma R, Omura S. J Antibiot. 1999;52:689–694. doi: 10.7164/antibiotics.52.689. [DOI] [PubMed] [Google Scholar]
- 23.Kasai Y, Komatsu K, Shigemori H, Tsuda M, Mikami Y, Kobayashi J. J Nat Prod. 2005;68:777–779. doi: 10.1021/np050046b. [DOI] [PubMed] [Google Scholar]
- 24.Wagner T, Fischer M. Mycol Prog. 2002;1:93–104. [Google Scholar]
- 25.Zhang N, Castlebury LA, Miller AN, Huhndorf SM, Schoch CL, Seifert KA, Rossman AY, Rogers JD, Kohlmeyer J, Volkmann-Kohlmeyer B, Sung GH. Mycologia. 2006;98:1076–1087. doi: 10.3852/mycologia.98.6.1076. [DOI] [PubMed] [Google Scholar]
- 26.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 27.Zwickl DJ. Ph.D. dissertation. The University of Texas; Austin: 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. [Google Scholar]
- 28.Page RDM. Computer Applications in the Biosciences. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.