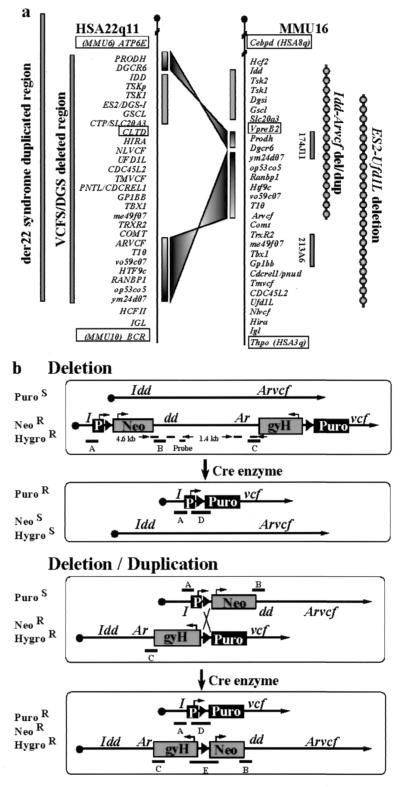
Figure 1.
(a) Comparison of the human VCFS/DGS region with the homologous region in mice. The relative order of genes on Homo sapiens chromosome 22q11 (HSA22q11) and Mus musculus chromosome 16 (MMU16) is shown (11, 18). The location of bacterial artificial chromosomes (BACs) 147J11 and 213A6 used for fluorescence in situ hybridization experiments and the region deleted in our experiments and those of Lindsay et al. (13) are shown on the right. Ym24do7, op53c05, me49f07, and vo59c07 correspond to independent genes identified by genscan analysis of GenBank sequences AC006082, AC003060, AC003066, AC008019, which are also represented in the 22q11 genomic sequence. (b) Scheme for selecting Cre/LoxP-mediated interstitial chromosomal deletion and duplication. Recombination between LoxP sites in cis leads to a deletion (upper two diagrams), and recombination between LoxP sites in trans leads to a deletion as well as a duplication (lower two diagrams). The locations of various diagnostic PCR products are represented by A–E. The Idd targeting vector contained a 4.3-kb XbaI genomic fragment including Idd exon 6, a 3.0-kb PvuII/SmaI genomic fragment including Idd exon 2, and part of exon 3 and a cassette containing a gene that confers resistance to neomycin (NeoR). The cassette also included a LoxP site and a solitary phosphoglycerate kinase (PGK) promoter (P). The Arvcf targeting vector contained a 0.7-kb EcoRI/MscI fragment containing a part of Arvcf exon 3 and an 8.0-kb XbaI fragment including Arvcf exons 5 and 6. A 3.1-kb cassette containing a hygromycin resistance (HygroR) gene, a LoxP site, and a promoterless puromycin resistance (PuroR) gene was inserted by blunt-end ligation into the MscI and XbaI sites.