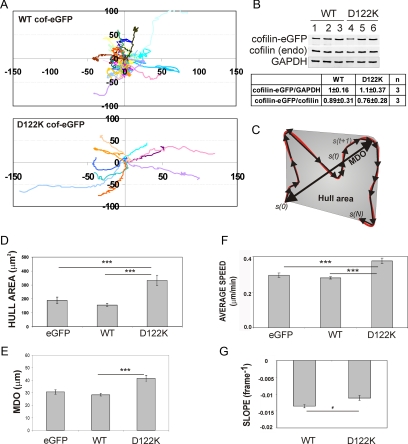
Figure 4.
D122K cofilin overexpression results in faster and more directional fibroblast migration. (A) Cell trajectories (a selection is shown of the cell populations considered) of NIH3T3, migrating on fibronectin and either overexpressing WT (top) or D122K cofilin-eGFP (bottom). The initial position of each track is superimposed on (0,0). (B) Quantification of expression levels of WT or D122K cofilin-eGFP and ratio of cofilin-eGFP to endogenous cofilin in transfected NIH3T3 cells. Western blot with anti-cofilin signal is shown for WT (lanes 1–3) and D122K (lanes 4–6) cofilin–eGFP-expressing cells (3 independent samples). The GAPDH signal is used to correct for equal loading on the gel. In comparing WT and D122K cofilin expression levels, the cofilin-eGFP/GADPH ratio was set to 1 for WT (C) Cartoon illustrating migration parameters: cell trajectory in red, separate migration steps in black. s(0), origin, s(N), end position of trajectory. MDO and Hull area (gray) are measures of distance and area covered by a cell, respectively (see Supplemental Data). Mean value for Hull area (D), mean MDO (E), and average speed (F) for a population of NIH3T3 cells expressing eGFP (n = 155 cells), overexpressing WT (n = 390) or D122K cofilin-eGFP (n = 140). (G) Mean SLOPE, a parameter for persistence (see Supplemental Data), for NIH3T3 cell populations overexpressing WT (n = 204) or D122K cofilin-eGFP (n = 71). A less negative SLOPE value reflects a higher directionality of cell movement. (D–G) Mean or average values ± SEM; statistical significance of differences between mean values are calculated using Mann–Whitney tests and indicated by *, p < 0.05 and ***, p < 0.001.