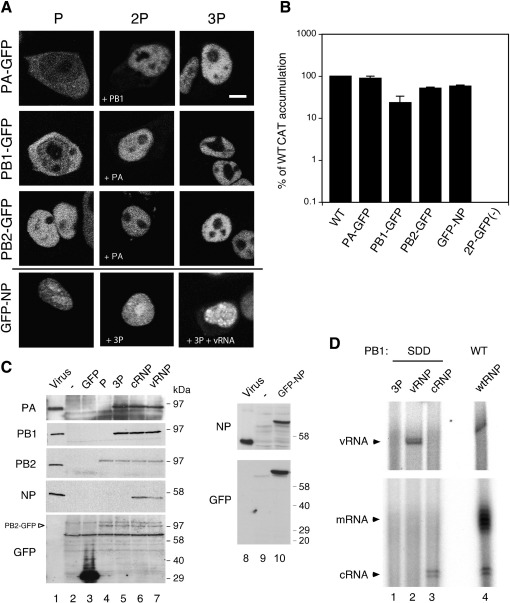
Fig. 1.
Reconstitution of recombinant GFP-tagged influenza RNPs in 293T cells. (A) Living cells expressing GFP-tagged RNP subunits alone or in combinations as indicated (2P; polymerase protein pairs, as labelled, 3P; the full polymerase complex) were analysed directly for GFP expression by confocal microscopy. Scale bar 5 μm. (B) Cells were transfected with plasmids expressing a synthetic vRNA encoding CAT and three (2P-GFP) or four of the RNP polypeptides, either all untagged (WT) or with the indicated one tagged with GFP. Three days post-transfection cells were analysed for CAT accumulation by ELISA. The mean and range from two independent experiments is plotted relative to the amount produced by WT RNPs. (C, D) Cells were transfected with plasmids expressing the indicated polypeptides (P, PB2-GFP; 3P, PB2-GFP, PA and PB1; cRNP and vRNP, PB2-GFP, PA and PB1 SDD along with NP and either a cRNA or vRNA containing a CAT gene respectively) or left untransfected (−). 24 h post-transfection (C) cell lysates and samples of purified virus (lane 1, as a marker) were analysed by SDS-PAGE and western blotting with antisera against PA, PB1, PB2, NP and GFP; (D) total RNA was extracted and analysed by primer extension using radiolabelled oligonucleotides specific for negative (top panel) or positive sense (lower panel) CAT transcripts. Radiolabelled products were separated by urea-PAGE and detected by autoradiography. Primer extension products resulting from the indicated CAT RNA species are labelled.