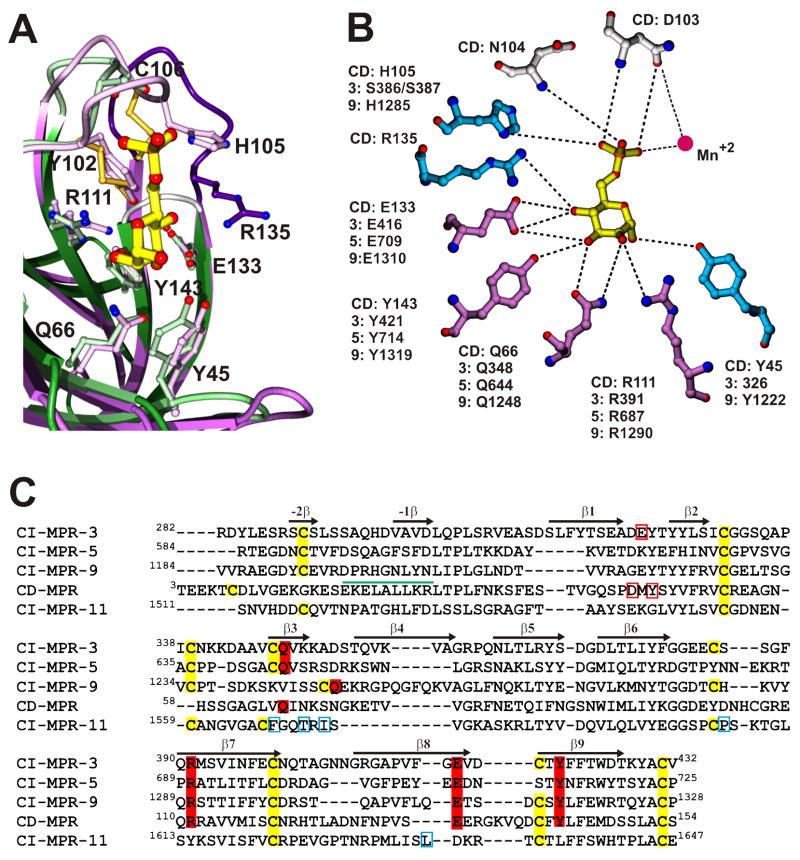
Figure 4.
Conservation of the Man-6-P binding site and essential residues for carbohydrate binding. A) Superposition of the Man-6-P binding sites of the CD-MPR (purple) and domain 3 of the CI-MPR (green). The architecture of both binding pockets is essentially the same with the exception of loop D (dark blue, CD-MPR; grey, domain 3), which is shorter in domain 3. B) A schematic drawing showing interactions between Man-6-P and residues in the CD-MPR and their homologous residues in domains 3, 5, and 9 of the CI-MPR. Dotted lines indicate potential hydrogen bonds. Mutational studies have shown that the four residues shown in purple are essential for Man-6-P binding and that mutation of the residues shown in blue partially decreased Man-6-P binding affinity. The two residues in grey have not been tested. C) Sequence alignment of domains 3, 5, 9 and 11 of the CI-MPR and the extracytosolic domain of the CD-MPR. The conserved cysteines are highlighted in yellow and the four residues that are essential for Man-6-P binding are highlighted in red. Residues that are within hydrogen bonding distance of Man-6-P in the crystal structures of CD-MPR and domains 1–3 of the CI-MPR, but found to be not essential for Man-6-P binding are boxed in red. Secondary structural elements are indicated with arrows and the single α-helix found in the CD-MPR is indicated with a green cylinder. Residues involved in IGF-II binding in domain 11 of the CI-MPR are boxed in blue.