Figure 2.
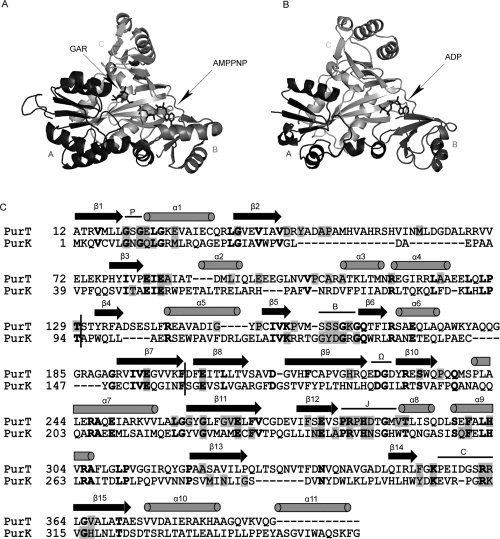
Three-dimensional structures of PurT with GAR and 5′-adenylyl imidodiphosphate (AMPPNP) (A), PurK with ADP (B), and a structure-based sequence alignment (C). In (A) and (B), each enzyme is divided into three structural motifs, the A-, B-, and C-domains which are color-coded in black, dark gray, and gray, respectively. The stick representation of the substrates in complex with enzymes is displayed in dark gray. The representations were prepared with Weblab Viewer Pro 3.7 (Molecular Simulations Inc.). In (C), a structure-based alignment was prepared using CE (combinatorial extension of the optimal path).19 The arrows and cylinders represent the β-sheet and α-helix secondary structures, respectively. Conserved residues in each enzyme are masked in gray and identical residues between PurT and PurK are shown bold. The vertical black bars label the boundaries of the A-, B-, and C-domains. Additionally, five conserved loop regions, the P-loop, B-loop, Ω-loop, J-loop, and C-loop, are labeled.
