Abstract
Metabolic syndrome, a clustering of conditions including obesity, insulin resistance, and hypertension, is a risk factor for cardiovascular morbidity and mortality. Because peroxisome proliferator-activated receptor γ (PPARγ) regulates adipocyte differentiation and lipid metabolism and is the molecular target of a class of insulin sensitizers, genetic variants that alter Pparg gene expression are potential contributors to the metabolic syndrome. To test this possibility, we generated mice having 182% of the normal steady-state level of PPARγ mRNA by replacing the 3′-UTR of the natural Pparg gene with that of the β-globin gene, thereby stabilizing the Pparg transcripts. This increase in PPARγ mRNA level had no apparent consequences in various physiological parameters, except that the mice repeatedly showed a trend toward lower blood pressures (by about 3 mm Hg) than their WT littermates. In contrast, the opposite trend, toward increased blood pressure, was observed in mice with genetically reduced levels of PPARγ mRNA as a consequence of insertion of an allele with an mRNA-destabilizing sequence into the endogenous 3′-UTR of the Pparg gene. By combining 12 sets of blood pressure measurements in more than 350 mutant mice having PPARγ expression levels varying from 28% to 182% and more than 280 WT littermates, we show that a 2-fold genetic increase (or decrease) in PPARγ expression levels decreases (or increases) blood pressure by about 2.8 mm Hg. Thus, our experiments demonstrate that quantitative variants causing decreased Pparg expression are a potential causative risk factor for essential hypertension.
Keywords: 3′-UTR, hypertension, metabolic syndrome, mouse models
Awareness is increasing that a significant number of adults in industrialized countries develop a metabolic syndrome characterized by a clustering of several abnormal traits, including insulin resistance, dyslipidemia, essential hypertension, visceral obesity, glucose intolerance, and noninsulin-dependent diabetes mellitus (1). Although a familial tendency toward metabolic syndrome suggests a genetic risk component, loci that predispose individuals to this syndrome remain largely undefined. This is partly because susceptibility to these conditions is likely determined by combinations of small changes in many genes. In addition, although numerous genome-wide studies have been carried out to identify single-nucleotide polymorphisms in the human genome, determining whether an “association” identified by epidemiologic studies is “causative” remains a great challenge. This problem is particularly evident with genetic variations in noncoding sequences. Nevertheless, the problem of determining whether quantitative genetic differences cause complex changes can be resolved by the use of animal models, as we report here for peroxisome proliferator–activated receptor γ (PPARγ).
PPARγ is a nuclear receptor that plays important roles in the regulation of adipocyte differentiation and glucose homeostasis, and also mediates the insulin-sensitizing effects of a class of drugs, the thiazolidinediones (TZDs). Consequently, the gene coding for PPARγ (Pparg) is an excellent candidate for playing a causative role in the development of metabolic syndrome. Several human PPARγ variants, spanning a wide spectrum of activity, have been identified; however, the relationships between these inherited differences in PPARγ activity and the traits associated with the human metabolic syndrome remain unclear. For example, the polymorphic P12A substitution, which leads to a lower affinity for the peroxisome proliferator response element and decreased transactivation in in vitro studies (2, 3), is associated with lower body mass index and enhanced insulin sensitivity in some, although not all, population studies (2, 4). In contrast, 4 unrelated individuals carrying a P115Q substitution, which increases PPARγ activity by preventing S114 phosphorylation, were severely obese and appeared to have greater-than-expected insulin sensitivity (5). On the other hand, another individual carrying the P115Q mutation was moderately overweight and had decreased insulin sensitivity (6). Other PPARγ mutations that result in production of transactivation-defective proteins and decreased PPARγ activity, such as P467L, V290M, F360L, and R397C, are associated with lipodystrophy and severe insulin resistance (7).
Extensive work exploring the influence of PPARγ variations on metabolism has been done using mouse models. A complete lack of PPARγ is embryonically lethal in homozygous knockout mice (8, 9). Heterozygous PPARγ-deficient mice are protected from high-fat diet–induced weight gain and loss of insulin sensitivity (9). Mice homozygous for a constitutively active form of PPARγ resulting from an S112A mutation (mouse S112 is equivalent to human S114) gain weight normally and are protected against obesity-induced insulin resistance (10). Mice with a mutation equivalent to the human P467L mutation maintain normal insulin sensitivity with altered body fat distribution and mild hypertension (11). The metabolic effects of simple increases or decreases in PPARγ expression level, such as can occur in humans as a consequence of normal variations in noncoding or regulatory regions of the gene, have not yet been explored.
In a previous study, we showed that the steady-state expression of a gene can be modulated in vitro at the mRNA and protein levels by changing its 3′-UTR, with minimal effects on the endogenous regulating machinery and no changes in the coding sequence (12). In the present study, we used this approach to study the in vivo effects of altering the steady-state expression level of PPARγ by changing the 3′-UTR of the Pparg gene in mice.
Results
Mice With Increased Pparg Expression.
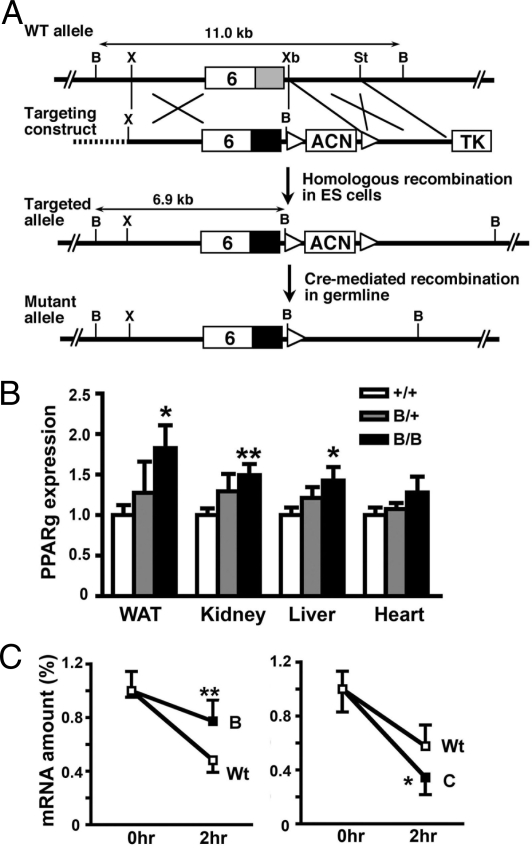
The targeting strategy used to replace the WT mouse PPARγ 3′-UTR (the WT allele) with the β-globin 3′-UTR (the B allele) is illustrated in Fig. 1A. The resulting modified Pparg-B allele retains all of the normal PPARγ regulatory elements in the promoter and coding sequences.
Fig. 1.
Mice with increased PPARγ gene expression. (A) Gene-targeting strategy. The WT mouse Pparg locus has a 345-bp 3′-UTR (gray box) in exon 6. The targeting construct contains an 855-bp β-globin 3′-UTR (black box), ACN (testis-specific Ace promoter–driven Cre and pMC1Neo) cassette flanked by loxP sequences (white triangles), thymidine kinase (TK), and a plasmid vector sequence (dotted line). Homologous recombination in ES cells results in the targeted allele having a 6.9-kb BamHI fragment instead of an 11.0-kb endogenous fragment that hybridized to a probe from the exon 6 coding region. The ACN cassette is excised on germline transmission, and subsequent PpargB/+ offspring carry a mutant allele. B, X, Xb, and St indicate BamHI, XhoI, XbaI, and StuI restriction enzyme sites, respectively. (B) PPARγ mRNA levels in PpargB/+ (n = 7) and PpargB/B (n = 9) tissues relative to those of Pparg+/+ (n = 10) tissues. *, P < .05; **, P < .01 against Pparg+/+ littermates. (C) PPARγ mRNA stability. Hepatic mRNA was isolated from heterozygous PpargB/+ mice (Left Panel) or PpargC/+ mice (Right Panel) before and 2 h after a tail vein injection of actinomycin D and α-amanitin. Between 6 and 8 mice were used for each time point. The levels of PPARγ mRNA were determined by allele-specific RT-PCR and are expressed relative to the levels at the zero time point. *, P < .05; **, P < .001 between 2 two alleles. Wt, WT allele; B, Pparg-B allele; C, Ppar-C allele.
PpargB/+ heterozygotes produced offspring in the expected Mendelian proportions (Pparg+/+:PpargB/+:PpargB/B = 68:127:48; P = .85 by the χ2 test). Quantitative RT-PCR showed that the steady-state PPARγ message level in PpargB/B mice was 183% of that in WT littermates in adipose tissue, 149% in the kidneys, 143% in the liver, and 128% in the heart (Fig. 1B). The corresponding PPARγ message levels in heterozygous PpargB/+ mice were 128%, 129%, 121%, and 108%.
To confirm that the increased PPARγ expression is due to increased stability of the message, we measured the decay of the PPARγ transcripts from the Pparg-B allele relative to that from the WT allele in the livers of PpargB/+ heterozygotes after injection of actinomycin D. Real-time RT-PCR using allele-specific primers/probes showed that the PPARγ transcripts from the Pparg-B allele were more stable than those from the WT allele (down to ≈70% after 2 h vs ≈50% after 2 h) (Fig. 1C, Left Panel). This increased stability is in marked contrast to the mRNA in the livers of PpargC/+ heterozygotes, which have a Pparg-C allele with a mRNA destabilizing sequence from the c-fos gene inserted into its 3′-UTR region (13); the unstable transcript of Pparg-C allele was down to ≈30% after 2 h, whereas mRNA from the WT allele was ≈55% (Fig. 1C, Right Panel).
Normal Metabolic Profiles in Mice with the Pparg-B Allele.
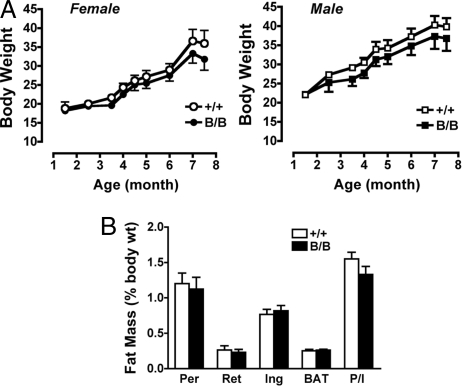
The homozygous PpargB/B mice developed normally and were healthy. No abnormalities were found in the mutant mice externally or internally on macroscopic examination. The growth curves of both male and female PpargB/B mice were the same as those of WT littermates fed regular chow. High-fat feeding increased the body weight of all of the mice regardless of genotype (Fig. 2A). Metabolic parameters from a set of PpargB/B mice and their littermates are compared in Table S1. The organ weights and total weights, as well as individual fat pad weights [perigonadal, retroperitoneal, and inguinal white adipose tissue (WAT) and interscapular brown adipose tissue] did not differ significantly between the PpargB/B and WT mice, and the body fat distribution was not altered in the PpargB/B mice (Fig. 2B). Fasting plasma total cholesterol and free fatty acids also were not altered in the PpargB/B mice (Table S1). In this set of littermates, plasma levels of triglycerides were modestly but significantly lower in the PpargB/B mice compared with the WT mice in both females (30 ± 1 mg/dL vs. 42 ± 3 mg/dL) and males (35 ± 1 mg/dL vs. 41 ± 3 mg/dL) (P < .002 by ANOVA). However, plasma levels of triglycerides were not significantly different in the 3 other littermate groups (overall P = .7 by ANOVA, with litter group, sex, and genotype as factors; n = 45 for PpargB/B and n = 58 for Pparg+/+).
Fig. 2.
Body weights and body fat distribution. (A) Body weight increases in females (Left Panel: Pparg+/+, n = 7; PpargB/B, n = 5) and males (Right Panel: Pparg+/+, n = 10; PpargB/B, n = 7) fed a high-fat diet. No statistical differences were noted at any time point. (B) Adipose tissue mass in 14-week-old male Pparg+/+ (n = 10) and PpargB/B (n = 9) mice fed regular chow, expressed as percentage of body weight. Per, Ret, and Ing represent perigonadal, retroperitoneal, and inguinal WAT, respectively; BAT indicates interscapular brown adipose tissue; P/I indicates the ratio of perigonadal fat weight to inguinal fat weight.
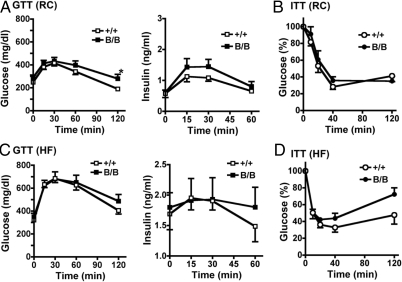
Plasma glucose and insulin concentrations were normal in PpargB/B mice in the fasted state. However, these mice exhibited slower glucose clearance than their WT littermates following a glucose load (Fig. 3A), as well as significantly higher plasma glucose levels 120 min after a glucose load (279 ± 38 mg/dL vs. 190 ± 17 mg/dL; P < .05). Insulin levels during glucose tolerance testing were not significantly different in the 2 genotypes. Insulin tolerance tests showed similar glucose-lowering effects of insulin in the 2 genotypes (Fig. 3B). Thus, the PpargB/B mice appeared to have normal insulin sensitivity but possibly slightly impaired postprandial glucose tolerance.
Fig. 3.
Normal glucose tolerance and insulin sensitivity in PpargB/B mice. (A) Plasma glucose (Left Panel) and insulin levels (Right Panel) during glucose tolerance tests on male mice fed regular chow (14≈16 weeks old; n = 8–10). (B) Insulin tolerance tests on female mice fed regular chow (14≈16 weeks old; n = 7–12). Data are expressed as percentage of plasma glucose level before insulin injection. (C) Glucose tolerance tests on males fed a high-fat diet for 3 months (20∼22 weeks old; n = 14–19). Plasma levels of glucose (Right Panel) and insulin (Left Panel) are shown. (D) Insulin tolerance tests on females on a high-fat diet for 3 months (20∼22 weeks old; n = 12–16). *, P < .05 against Pparg+/+ littermates.
We next examined the effect of a high-fat diet on the insulin sensitivity of these animals. Glucose clearance and insulin secretion following a glucose load were similar in the PpargB/B mice and their WT littermates fed a high-fat diet for 3 months (Fig. 3C). The PpargB/B mice showed a decreased hypoglycemic effect in response to exogenous insulin compared with their WT littermates, although the differences in plasma glucose level at specific time points were not significant (Fig. 3D). Taken together, these results indicate that an increase in the expression of the Pparg gene to 182% of normal has only a minor impact on glucose metabolism, but could lead to a decrease in insulin sensitivity when combined with a high-fat diet.
PPARγ Gene Expression and Blood Pressure.
Blood pressure assessed in conscious mice by tail-cuff measurement showed that both 4-month-old male and female PpargB/B mice had lower blood pressure (by ≈3 mm Hg) than their WT littermates (genotype effect, P = .09 by ANOVA; Table 1). Pulse rates did not differ in these 2 genotypes in either males or females. Blood pressures were also lower in PpargB/B mice fed a high-fat diet for 4 months compared with their WT littermates (genotype effects, P = .01 and .06 by ANOVA, respectively; Table 1). Although the differences in average blood pressure of the PpargB/B and Pparg+/+ mice in the 6 groups of animals from the 3 experiments did not reach statistical significance, it is noteworthy that the PpargB/B mice had lower blood pressures than their WT littermates in all 6 groups of animals that differed by diet, age, and gender.
Table 1.
Tail-cuff blood pressure measurements in PpargB/B (B/B) and Pparg+/+(+/+) mice
| Experiment | Females |
Males |
P | ||
|---|---|---|---|---|---|
| B/B | +/+ | B/B | +/+ | ||
| Blood pressure, mm Hg | |||||
| [a] 4 months, RC | 100 ± 2 (13) | 103 ± 2 (18) | 97 ± 2 (15) | 100 ± 2 (18) | .09 |
| [b] 8 months, HF | 100 ± 2 (13) | 103 ± 2 (19) | 94 ± 3 (13) | 100 ± 1 (18) | .01 |
| [c] 6 months, HF | 94 ± 2 (10) | 96 ± 2 (15) | 93 ± 1 (10) | 98 ± 4 (6) | .06 |
| Pulse rate, beats/min | |||||
| [a] 4 months, RC | 734 ± 8 (13) | 735 ± 7 (17) | 724 ± 16 (13) | 716 ± 14 (18) | .70 |
| [b] 8 months, HF | 720 ± 9 (13) | 740 ± 8 (18) | 699 ± 8 (12) | 719 ± 7 (18) | .02 |
| [c] 6 months, HF | 680 ± 11 (10) | 688 ± 8 (15) | 653 ± 9 (10) | 640 ± 15 (6) | .80 |
RC, regular chow; HF, Western-style high-fat diet. Data are mean ± SEM. The numbers of mice are given in parentheses. [a], [b], and [c] indicate experiment numbers. P values show genotype effects by ANOVA, with gender and genotype as 2 factors.
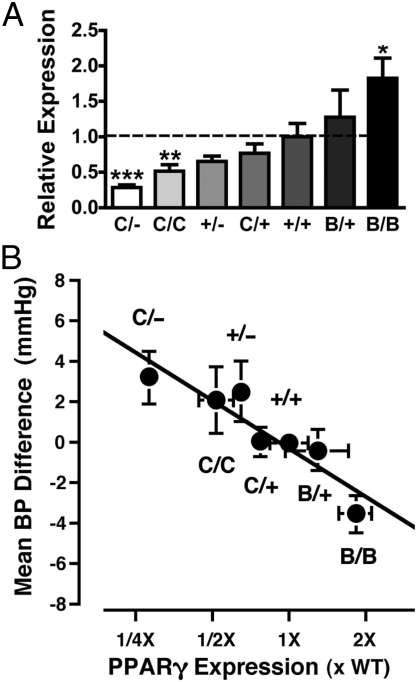
This trend toward reduced blood pressures in mice with 182% of normal PPARγ expression encouraged us to examine the blood pressure effects of PPARγ gene expression levels over a wider range. Seven genotypes of mice were used; their PPARγ expression levels relative to WT mice (100%) in WAT, in ascending order, were PpargC/−, 28%; PpargC/C, 52%; Pparg+/−, 60%; PpargC/+, 77%; Pparg+/+, 100%; PpargB/+, 128%; and PpargB/B, 182% (Fig. 4A). We observed a trend toward higher blood pressures in mice with reduced PPARγ gene expression; blood pressure was higher in the PpargC/− mice than in their Pparg+/+ littermates in 5 of 6 sets, analyzing males females separately, and the differences were significant in 2 sets (Table S2C).
Fig. 4.
Effects of increased/decreased PPARγ gene expression on blood pressure. (A) Average PPARγ mRNA levels in WATs of PpargC/− (C/−), PpargC/C (C/C), Pparg+/− (+/−), PpargC/+ (C/+), PpargB/+ (B/+), and PpargB/B (B/B) mice relative to their Pparg+/+ littermates (+/+, 100%). *, P < .05; **, P < .01; ***, P < .001 against Pparg+/+ mice. There were 7–9 mice in each group. (B) Mean ± SEM of the blood pressure differences in individual mice from the mean blood pressure of Pparg+/+ littermates within each experiment. n = 49, 40, 38, 115, 285, 51, and 73 for C/−, C/C, +/−, C/+, +/+, B/+ and B/B mice, respectively. The x-axis is the fold change in the average level of PPARγ gene expression in each genotype group from that of their WT littermates determined in (A). The black line indicates a slope of 2.8 mm Hg per 2-fold difference in the expression of PPARγ. The correlation between the mean blood pressure difference for each genotype and the mean expression level for each genotype was r = −0.92 (P < .005; n = 7).
In 12 experiments, designated [a]–[l], carried out over a 3-year period, blood pressures were measured in a total of 356 mutants and their 285 WT littermates (Tables 1 and S2). Each experiment involved litters matched with respect to age, diet, and genetic background. Because these factors differed between the 12 experiments, but not within each experiment, we analyzed the effects of PPARγ expression on blood pressure treating the experiment as a random effect. The analysis showed that blood pressure was significantly influenced by the mean level of PPARγ mRNA in mice of the 7 PPARγ genotypes studied (P < .0012; parameter estimate of −2.79 ± 0.86 mm Hg per 2-fold difference in expression; Table S3). This relationship is illustrated in Fig. 4B, in which the difference in blood pressure of individual animals from the mean blood pressure of the WT littermates within each experiment was used to derive an average blood pressure difference from normal for each genotype group. This difference was then plotted against the average fold change in PPARγ mRNA level for mice of the corresponding genotype. The combined data clearly indicate an inverse relationship between PPARγ expression level and blood pressure, with a 2-fold change in the level of PPARγ mRNA altering blood pressure by about 2.8 mm Hg.
Discussion
Taking advantage of the mRNA stabilizing/destabilizing features of the 3′-UTR sequence, we created a Pparg-B allele that confers an inherited increase in the level of PPARγ gene expression in mice without directly affecting the transcriptional regulation of the gene. Transcripts from the Pparg-B allele retain the original PPARγ coding sequences but also have a 3′-UTR from a β-globin gene. Our expectation that the steady-state expression level of the PPARγ gene would increase in mice carrying the Pparg-B allele was confirmed by the level of PPARγ message, which in PpargB/B homozygotes was 182% of that in WT littermates. Remarkably, this expression change occurred not only in adipose tissues, where PPARγ expression is high, but also in the liver and kidney, where PPARγ expression is much lower. The availability of the Pparg-B allele, together with the Pparg-C allele (which has mRNA- destabilizing elements of the c-fos gene inserted into the endogenous Pparg 3′-UTR), allowed us to genetically vary PPARγ expression between 28% and 182%.
The half-life of PPARγ mRNA in a tissue culture system has been reported to be ≈2.8 h (14), which is similar to our observation of ≈2 h in vivo in the liver. Some 3′-UTR sequences, such as that of β-globin mRNA (half-life ≈17 h), are known to have message-stabilizing properties (15), while other 3′-UTR sequences, such as that in c-fos mRNA (half-life ≈0.3 h), lead to very unstable mRNA (16). In our study, the half-life of the PPARγ message was approximately doubled in the Pparg-B allele and approximately halved in the Pparg-C allele. Thus, the observed alteration in the steady-state levels of the PPARγ message in mice carrying these Pparg alleles is a direct consequence of changes in mRNA stability in vivo. We note that the range of variations in the steady-state level of PPARγ mRNA (52%–182% of normal) resulting from the 3′-UTR modifications is significantly smaller than that expected from tests with a reporter gene expression system in cultured cells using the same 3′-UTR sequences (10%–350% of normal) (12). But in the reporter expression system, the GFP gene with a test 3′-UTR sequence is inserted into the Hprt locus and is driven by a constitutive β-actin promoter, and consequently lacks the endogenous feedback regulation and chromosomal context of the native gene. Furthermore, factors that affect message stability likely differ between tissues in vivo and embryonic stem cells in culture. Moreover, we cannot exclude the possibility that the altered 3′-UTR sequences in the endogenous Pparg gene, in addition to their posttranscriptional effects, might have affected chromatin remodeling and transcription levels. Nevertheless, the changes in the Pparg gene expression observed in our mutant animals are within the physiological range that might occur in humans.
While activation of PPARγ by TZD in both humans and mice clearly increases insulin sensitivity and increases body and fat weights (17), the nearly double PPARγ message seen in the homozygous PpargB/B mice did not affect their body or total fat weights. In addition, the PpargB/B mice had normal insulin sensitivity. The absence of any significant effect on these variables of doubling PPARγ mRNA levels could be due to the existence of posttranscriptional compensation, although protein synthesis usually increases or decreases roughly in proportion to the amount of RNA transcripts. Alternatively, moderate overexpression of PPARγ by itself may not be sufficient to change the phenotypic outcome. This possibility also is suggested by studies in cultured cells showing that overexpression of WT PPARγ has only a weak adipogenic effect in the absence of ligands (18, 19).
Despite these various caveats, our results demonstrate that modestly increased PPARγ mRNA levels lower basal blood pressure. The effect is small, and a large number of experimental animals with both higher-than-normal and lower-than-normal Pparg expression were needed to reach this conclusion. Nonetheless, we stress that each set of blood pressure measurements was carried out with littermates (mutant and WT), which allowed us to estimate that a 2-fold genetic increase (or decrease) in PPARγ expression decreases (or increases) blood pressure by about 2.8 mm Hg. Most of our current knowledge concerning the blood pressure–decreasing effects of increased PPARγ activity is based on the effect of TZDs (17). This relationship was first observed in patients with type 2 diabetes and was found to be independent of changes in insulin sensitivity. TZDs also have been found to attenuate the blood pressure increase resulting from angiotensin II infusion in rats (20) and to improve vascular function in normoglycemic mice with hypertension (21). Data on the effects of decreasing PPARγ activity are available as well. Thus, humans with the dominant-negative P467L mutation are hypertensive, and we have previously shown that an equivalent PPARγ-P465L mutation in mice causes a ≈7 mm Hg increase in blood pressure (11). Transgenic mice that overexpress the human PPARγ gene with a P467L mutation in vascular smooth muscle cells also have elevated blood pressure (22). Our demonstration of an inverse relationship between genetically determined levels of PPARγ expression and blood pressure is consistent with these observations.
The increased blood pressure in our mice with decreased Pparg expression differs from the hypotensive phenotype described in mice with a generalized lack of PPARγ (23). A complete absence of PPARγ causes embryonic death, but the investigators in that previous study were able to rescue them from lethality by using a Cre-loxP system that deletes the Pparg gene in the epiblast (and so also in the embryos), but not in extraembryonic tissues. The rescued PPARγ-null mice, designated MORE-PGKO, were severely lipodystrophic and insulin-resistant, but had decreased blood pressure as measured by telemetric recording in a limited number of mice. In contrast, the mutants generated in our experiment had no overt metabolic phenotype, except that the PpargC/− mice with 28% normal expression had ≈30% less fat mass and were mildly insulin-resistant (24), and in our study blood pressure measurement was done using the tail-cuff method in a large number of mice. The different outcomes of the 2 experiments could be related to the difference between the effects of a modest quantitative reduction in the expression of a protein compared with the effects of the complete absence of that protein. Further studies are needed to elucidate the precise mechanisms through which PPARγ expression regulates blood pressure.
The changes in the PPARγ expression levels in the series of Pparg mutants that we have generated and studied are within a physiological range that may occur in humans as a consequence of polymorphic variations. Moreover, these mice had no changes in the genome other than in the Pparg 3′-UTR sequence, and they produced normal PPARγ protein in all relevant tissues. Consequently, these mice are valuable predictors of the effects of quantitative genetic variations in human individuals. The clear inverse relationship between basal expression levels of PPARγ and blood pressure revealed in our experiments in mice suggests that variations in the Pparg locus could play a causative role in determining basal blood pressures in humans. Thus, although the 2- to 3-mm Hg change in the basal blood pressure that we observed when PPARγ expression was doubled or halved is not large, it is comparable to that seen in other human polymorphisms. For example, the M235T polymorphism in the angiotensinogen gene in humans is in linkage disequilibrium with a promoter variant A-6T that causes a 2-fold difference in expression level (25). A recent meta-analysis involving 26,818 subjects from 46 studies reported a small but significant association of the 235T allele with hypertension, with an odds ratio of ≈1.2 (26). In a large, homogeneous study population, Ortlepp et al. (27) estimated that the M235T polymorphism explains about 2 mm Hg of the difference in diastolic blood pressure. The difficulty in establishing the existence of such small effects may be partly responsible for the lack of agreement between published studies of genetic associations between Pparg polymorphisms and hypertension in humans (28–30). Our study of mice with varying Pparg expression created by modifying its 3′-UTR sequence indicates that quantitative variants causing decreased Pparg expression are a potential causative risk factor for essential hypertension.
Materials and Methods
Targeting Construct Design.
A mouse genomic DNA (129/SvEv strain) containing PPARγ exon 6 and 3′-UTR was cloned from a λ phage library, and 5.5-kb and 9.4-kb XhoI fragments were subcloned into pBlueScript. To generate mice with increased steady-state levels of PPARγ mRNA, a 345-bp BglII/NdeI fragment containing the PPARγ 3′-UTR was replaced with an 855-bp rabbit β-globin 3′-UTR (Fig. 1A). The fragment was cloned into the targeting vector, which contains a 6.5-kb XhoI/XbaI 5′-homology fragment, the ACN (testis-specific ACE promoter, Cre recombinase, and neomycin- resistance gene) cassette (31), a 1.3-kb XbaI/StuI 3′-homology fragment, and a thymidine kinase gene.
Generation of PPARγ Mutant Mice.
Electroporation of linearized vector into TC-1 ES cells of 129S6 origin and selection with G418 and ganciclovir were performed as described previously (32). Successful modifications of the Pparg locus were identified by PCR and confirmed by Southern blot analysis, which showed the presence of a 6.9-kb BamHI restriction fragment expected for the Pparg-B allele when hybridized with a probe derived from the exon 6 sequence. Targeted ES cell clones were injected into C57BL/6 blastocysts and transferred into pseudopregnant females to obtain chimeric offspring. The ACN cassette, flanked by loxP sequences, was excised out in mice on germline transmission. Male chimeras transmitted the mutant PPARγ alleles, and the lines were maintained on 129S6 genetic background. The Pparg-B allele was placed onto C57BL/6J by backcrossing for 4–5 generations before intercrossing to generate Pparg+/+, PpargB/+, and PpargB/B littermates for subsequent studies. The production of mice carrying the Pparg-C allele with an mRNA-destabilizing sequence from the c-Fos gene inserted into its 3′-UTR region was described previously (13, 24). The mice were housed in a pathogen-free barrier facility (12 h/12 h light/dark cycle) and fed ad libitum either a regular chow diet (LabDiet 5P76; PMI Nutrition International) or a high-fat diet (D12451; Research Diets) consisting of 45% calories as fat, 35% as carbohydrate, and 20% as protein. All animal studies were approved by the Institutional Animal Care and Use Committees of the University of North Carolina-Chapel Hill and the National Cheng Kung University.
Tissue Collection and RNA Analysis.
Mice were euthanized with an overdose of 2,2,2,-tribromoethanol, and tissues were collected and stored in RNAlater (Ambion). Tissue RNA was extracted using the RNAeasy Kit (Qiagen), and mRNA was analyzed by real-time quantitative RT-PCR using an ABI Prism 7700 (Applied Biosystems), with β-actin or Gapdh as the reference gene in each reaction. To assay RNA degradation, heterozygous mice were injected with actinomycin D (1.5 μg/g body weight; Calbiochem) and α-amanitin (0.5 μg/g body weight; Sigma) through the tail vein. Liver mRNA was isolated before and 2 h after injection, and the message level for each allele was analyzed using allele-specific oligo primers.
Lipid Metabolism.
Plasma total cholesterol (Cholesterol E; Wako Chemicals), free fatty acid (NEFA C; Wako Chemicals), and triglyceride (Stanbio Laboratory) were measured using standard colorimetric tests.
Glucose and Insulin Tolerance Tests.
Mice were fasted for 5 h and injected i.p. with glucose (1 g/kg body weight) or human regular insulin (0.5 or 0.75 U/kg body weight; Humulin; Eli Lilly). Blood samples were collected before and at indicated times after injections. Plasma glucose concentration was determined by a glucose colorimetric test (Autokit Glucose; Wako Chemicals). Insulin concentration was measured with rat insulin ELISA (Crystal Chemical).
Tail-Cuff Blood Pressure Measurements.
Blood pressure and pulse rate were measured noninvasively in conscious, restrained mice by the tail-cuff method (33). The blood pressure of an individual animal was represented by the mean of the daily averages of 30 trials per day for 6 consecutive days. Each set of experiments included an average of 12 (at least 5) mice of each genotype and gender group and similar numbers of WT littermates. Experimental sets differed by age, diet, and genetic background of the mice. Blood pressures of PpargB/+ and PpargB/B littermates were from intercrosses of the fifth-generation backcross toward C57BL/6J, and those of PpargC/C and PpargC/+ mice were measured in F2 littermates between 129S6 and C57BL/6J. PpargC/− and Pparg+/− mice were F1 between PpargC/+ heterozygotes (at the seventh generation backcrossed to C57BL/6J) and Pparg+/− heterozygotes on 129S6 background (8), provided by Dr. Ronald Evans (Salk Institute).
Data Analysis.
Values are reported as mean ± SEM. Statistical analysis was conducted with 2-way ANOVA, using genotype and gender as factors. The Student t test was used for comparisons between mutant and WT mice within each group, and differences were considered statistically significant at P < .05. To estimate the effect of PPARγ expression on blood pressure, the mean mRNA level in the WAT of mice in each genotype determined in a separate set of animals was treated as continuous variable, using an ANCOVA model, treating the experiment as a random effect. The animals' age, sex, and diet were included in the model as fixed effects.
Supplementary Material
Acknowledgments.
We thank Dr. Xiaohai Wan for guidance with the statistical analysis; Drs. Kumar Pandya and Nobuyuki Takahashi for discussion and critical reading of the manuscript; and A. Pandse, J. Hagaman, and S. Jordon for technical assistance. This work was supported by grants from the National Institutes of Health (HL49277, to O.S. and HL77145 and DK67320, to N.M.) and the National Science Council of Taiwan (96–2320-B-006–048-MY3, to Y.S.T.).
Footnotes
The authors declare no conflicts of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909657106/DCSupplemental.
References
- 1.Grundy SM, Brewer HB, Jr, Cleeman JI, Smith SC, Jr, Lenfant C. Definition of metabolic syndrome: Report of the National Heart, Lung, and Blood Institute/American Heart Association conference on scientific issues related to definition. Circulation. 2004;109:433–438. doi: 10.1161/01.CIR.0000111245.75752.C6. [DOI] [PubMed] [Google Scholar]
- 2.Deeb SS, et al. A Pro12Ala substitution in PPARgamma2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nat Genet. 1998;20:284–287. doi: 10.1038/3099. [DOI] [PubMed] [Google Scholar]
- 3.Masugi J, Tamori Y, Mori H, Koike T, Kasuga M. Inhibitory effect of a proline-to-alanine substitution at codon 12 of peroxisome proliferator–activated receptor-gamma 2 on thiazolidinedione-induced adipogenesis. Biochem Biophys Res Commun. 2000;268:178–182. doi: 10.1006/bbrc.2000.2096. [DOI] [PubMed] [Google Scholar]
- 4.Doney A, et al. Haplotype analysis of the PPARgamma Pro12Ala and C1431T variants reveals opposing associations with body weight. BMC Genet. 2002;3:21. doi: 10.1186/1471-2156-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ristow M, Muller-Wieland D, Pfeiffer A, Krone W, Kahn CR. Obesity associated with a mutation in a genetic regulator of adipocyte differentiation. N Engl J Med. 1998;339:953–959. doi: 10.1056/NEJM199810013391403. [DOI] [PubMed] [Google Scholar]
- 6.Bluher M, Paschke R. Analysis of the relationship between PPAR-gamma 2 gene variants and severe insulin resistance in obese patients with impaired glucose tolerance. Exp Clin Endocrinol Diabetes. 2003;111:85–90. doi: 10.1055/s-2003-39235. [DOI] [PubMed] [Google Scholar]
- 7.Tsai YS, Maeda N. PPARgamma: A critical determinant of body fat distribution in humans and mice. Trends Cardiovasc Med. 2005;15:81–85. doi: 10.1016/j.tcm.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 8.Barak Y, et al. PPAR gamma is required for placental, cardiac, and adipose tissue development. Mol Cell. 1999;4:585–595. doi: 10.1016/s1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- 9.Kubota N, et al. PPAR gamma mediates high-fat diet–induced adipocyte hypertrophy and insulin resistance. Mol Cell. 1999;4:597–609. doi: 10.1016/s1097-2765(00)80210-5. [DOI] [PubMed] [Google Scholar]
- 10.Rangwala SM, et al. Genetic modulation of PPARgamma phosphorylation regulates insulin sensitivity. Dev Cell. 2003;5:657–663. doi: 10.1016/s1534-5807(03)00274-0. [DOI] [PubMed] [Google Scholar]
- 11.Tsai YS, et al. Hypertension and abnormal fat distribution but not insulin resistance in mice with P465L PPARgamma. J Clin Invest. 2004;114:240–249. doi: 10.1172/JCI20964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kakoki M, et al. Altering the expression in mice of genes by modifying their 3′ regions. Dev Cell. 2004;6:597–606. doi: 10.1016/s1534-5807(04)00094-2. [DOI] [PubMed] [Google Scholar]
- 13.Tsai YS, et al. A de novo deafwaddler mutation of Pmca2 arising in ES cells and hitchhiking with a targeted modification of the Pparg gene. Mamm Genome. 2006;17:716–722. doi: 10.1007/s00335-005-0191-z. [DOI] [PubMed] [Google Scholar]
- 14.Fu M, et al. Early stimulation and late inhibition of peroxisome proliferator–activated receptor gamma (PPAR gamma) gene expression by transforming growth factor beta in human aortic smooth muscle cells: Role of early growth-response factor-1 (Egr-1), activator protein 1 (AP1) and Smads. Biochem J. 2003;370:1019–1025. doi: 10.1042/BJ20021503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Russell JE, Liebhaber SA. The stability of human beta-globin mRNA is dependent on structural determinants positioned within its 3′ untranslated region. Blood. 1996;87:5314–5323. [PubMed] [Google Scholar]
- 16.Treisman R. Transient accumulation of c-fos RNA following serum stimulation requires a conserved 5′ element and c-fos 3′ sequences. Cell. 1985;42:889–902. doi: 10.1016/0092-8674(85)90285-5. [DOI] [PubMed] [Google Scholar]
- 17.Willson TM, Lambert MH, Kliewer SA. Peroxisome proliferator–activated receptor gamma and metabolic disease. Annu Rev Biochem. 2001;70:341–367. doi: 10.1146/annurev.biochem.70.1.341. [DOI] [PubMed] [Google Scholar]
- 18.Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 19.Shao D, et al. Interdomain communication regulating ligand binding by PPAR-gamma. Nature. 1998;396:377–380. doi: 10.1038/24634. [DOI] [PubMed] [Google Scholar]
- 20.Diep QN, et al. Structure, endothelial function, cell growth, and inflammation in blood vessels of angiotensin II–infused rats: Role of peroxisome proliferator–activated receptor-gamma. Circulation. 2002;105:2296–2302. doi: 10.1161/01.cir.0000016049.86468.23. [DOI] [PubMed] [Google Scholar]
- 21.Ryan MJ, Didion SP, Mathur S, Faraci FM, Sigmund CD. PPAR(gamma) agonist rosiglitazone improves vascular function and lowers blood pressure in hypertensive transgenic mice. Hypertension. 2004;43:661–666. doi: 10.1161/01.HYP.0000116303.71408.c2. [DOI] [PubMed] [Google Scholar]
- 22.Halabi CM, et al. Interference with PPARγ function in smooth muscle causes vascular dysfunction and hypertension. Cell Metab. 2008;7:215–226. doi: 10.1016/j.cmet.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duan SZ, et al. Hypotension, lipodystrophy, and insulin resistance in generalized PPARgamma-deficient mice rescued from embryonic lethality. J Clin Invest. 2007;117:812–822. doi: 10.1172/JCI28859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsai YS, et al. Decreased PPARγ expression compromises perigonadal-specific fat deposition and insulin sensitivity. Mol Endocrinol. 2009 doi: 10.1210/me.2009-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inoue I, et al. A nucleotide substitution in the promoter of human angiotensinogen is associated with essential hypertension and affects basal transcription in vitro. J Clin Invest. 1997;99:1786–1797. doi: 10.1172/JCI119343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pereira AC, et al. Antiotensinogen 235T allele “dosage” is associated with blood pressure phenotypes. Hypertension. 2003;41:25–30. doi: 10.1161/01.hyp.0000047465.97065.15. [DOI] [PubMed] [Google Scholar]
- 27.Ortlepp JR, et al. Relation between the angiotensinogen (AGT) M235T gene polymorphism and blood pressure in a large, homogeneous study population. J Hum Hypertens. 2003;17:555–559. doi: 10.1038/sj.jhh.1001587. [DOI] [PubMed] [Google Scholar]
- 28.Hasstedt SJ, Ren QF, Teng K, Elbein SC. Effect of the peroxisome proliferator–activated receptor-gamma 2 pro(12)ala variant on obesity, glucose homeostasis, and blood pressure in members of familial type 2 diabetic kindreds. J Clin Endocrinol Metab. 2001;86:536–541. doi: 10.1210/jcem.86.2.7205. [DOI] [PubMed] [Google Scholar]
- 29.Rodriguez-Esparragon FJ, Rodriguez-Perez JC, Macias-Reyes A, Alamo-Santana F. Peroxisome proliferator–activated receptor-gamma2-Pro12Ala and endothelial nitric oxide synthase-4a/bgene polymorphisms are associated with essential hypertension. J Hypertens. 2003;2:1649–1655. doi: 10.1097/01.hjh.0000084719.53355.20. [DOI] [PubMed] [Google Scholar]
- 30.Gallicchio L, et al. Genetic polymorphisms of peroxisome proliferator–activated receptors and the risk of cardiovascular morbidity and mortality in a community-based cohort in Washington County, Maryland. PPAR Res. 2008;2008:276581. doi: 10.1155/2008/276581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bunting M, Bernstein KE, Greer JM, Capecchi MR, Thomas KR. Targeting genes for self-excision in the germ line. Genes Dev. 1999;13:1524–1528. doi: 10.1101/gad.13.12.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piedrahita JA, Zhang SH, Hagaman JR, Oliver PM, Maeda N. Generation of mice carrying a mutant apolipoprotein E gene inactivated by gene targeting in embryonic stem cells. Proc Natl Acad Sci USA. 1992;89:4471–4475. doi: 10.1073/pnas.89.10.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krege JH, Hodgin JB, Hagaman JR, Smithies O. A noninvasive computerized tail-cuff system for measuring blood pressure in mice. Hypertension. 1995;25:1111–1116. doi: 10.1161/01.hyp.25.5.1111. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.