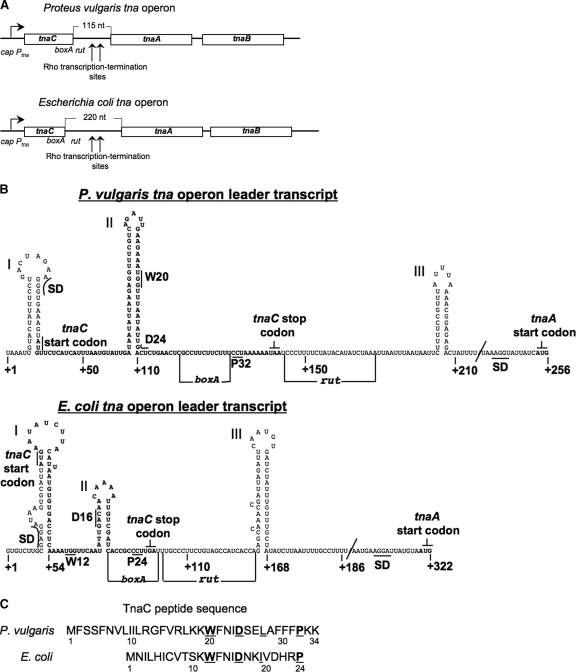
FIG. 1.
Comparison of the organizational features of the tna operon of P. vulgaris and of E. coli with special emphasis on their leader regulatory regions. (A) Schematic representation and comparison of the tna operon of P. vulgaris with that of E. coli. Boxes represent the three open reading frames within each operon. The arrow at the left indicates the direction of transcription initiated at the promoter/cap site of the tna operon. Transcription of these operons is modulated by catabolic repression involving interaction of the catabolite repressor protein with the cap site (15, 18). Vertical arrows indicate the operon regions where transcription is terminated prematurely under the influence of the Rho protein and the boxA and rut sequences. The lengths of the spacer regions separating the tnaC and tnaA genes are indicated. Numbers identify nucleotide positions in the mRNAs. (B) Schematic representation of the predicted secondary structure of the mRNA leader RNAs of both tna operons, from the nucleotide at position +1 to the G nucleotide of the tnaA start codon. Letters in boldface type indicate the nucleotides corresponding to the tnaC open reading frame. The positions of the Shine-Dalgarno (SD) sequence and start codon for the tnaC and tnaA genes are indicated. Some codons analyzed in this study as well as the stop codon of tnaC are also shown. The RNA segments boxA and rut involved in transcription termination induced by Rho factor are identified by horizontal lines. The structures were obtained using the Mfold web server (20). (C) Amino acid sequence of the TnaC peptides encoded by the tna operons. The letters in boldface type identify residues shown to be essential for induction. Numbers indicate residue positions in the peptide.