Lipoic acid (Fig. 1) is a cofactor that is critical to the catalytic function of several key enzymes in intermediary metabolism that are found in all domains of life. There are two pathways to lipoyl-modified proteins. The de novo pathway from octanoyl-acyl carrier protein (ACP) involves the consecutive action of LipB and LipA (Fig. 2A), and the LplA-dependent salvage pathway activates and transfers environmental lipoic acid to the lipoyl protein domains (Fig. 2B). In this issue, Hermes and Cronan (6) identify gain-of-function mutations in LplA that allow cells to bypass the requirement for LipB in de novo lipoic acid formation (Fig. 2C). Normally, lipB null strains require either a lipoate or an octanoate supplement for growth, but these lplA mutants have increased affinities for octanoic acid, permitting them to utilize the low concentrations of intracellular octanoate, thus substituting for the LipB reaction. The characterization of these suppressor strains provides new information on the biochemistry of lipoate metabolism and reveals the existence of intracellular pools of medium-chain fatty acids that previously were not thought to exist.
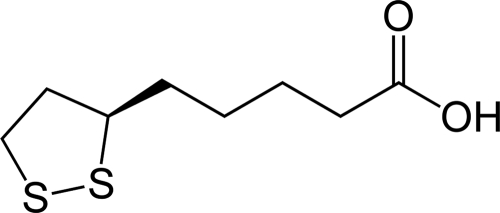
FIG. 1.
Structure of lipoic acid. Lipoic acid [R-5-(1,2-dithiolan- 3-yl)pentanoic acid] is sometimes known as thioctic acid.
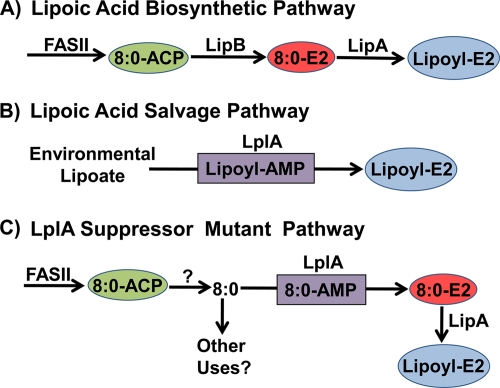
FIG. 2.
Lipoic acid biosynthesis in E. coli. (A) The de novo lipoate biosynthetic pathway begins by the utilization of octanoyl-ACP (8:0-ACP) produced by type II fatty acid synthase (FASII) by LipB to acylate the E2 domain of PDH. Octanoate bound to the E2 domain is converted to lipoic acid by the S-adenylmethionine-dependent LipA protein to form lipoyl-E2. (B) The scavenging pathway utilizes LplA to transfer lipoic acid from the environment to the E2 domain via an acyl-adenylate intermediate. (C) The LplA suppressor pathway is possible due to an intracellular pool of octanoate (8:0) released from the FASII pathway by an unknown thioesterase-like mechanism. Mutant LplA proteins with increased affinity for free octanoate activate and transfer the fatty acid to 8:0-E2. Lipoyl-E2 is then formed by LipA. It is not known whether there are other potential uses for the medium-chain free fatty acid pool in E. coli.
Functions of lipoic acid.
Lipoic acid is essential for the functions of several enzymes in oxidative metabolism (2). A good example is pyruvate dehydrogenase (PDH), where covalently bound lipoate shuttles reaction intermediates between the active sites of a multisubunit complex (11). Lipoic acid is bound to the dihydrolipoyl transacetylase (E2) subunit in its oxidized state as a five-membered, disulfide-linked ring (Fig. 1). The E1 subunit of PDH decarboxylates pyruvate, forming 2-hydroxyethyl-2-thiamine pyrophosphate, and the lipoic acid disulfide is reduced when E2 accepts an acetaldehyde moiety from the thiamine adduct. Acetyl-coenzyme A is then released via a thioester exchange reaction with coenzyme A. The disulfide ring of lipoate is reformed by the FAD-dependent dihydrolipoyl dehydrogenase component (E3). Other important bacterial lipoyl enzymes include the glycine cleavage system, α-ketoglutarate dehydrogenase, and the branched-chain α- ketoacid dehydrogenase that is involved in supplying branched-chain precursors for fatty acid synthesis. In Escherichia coli, strains defective in lipoic acid synthesis are unable to grow aerobically on glucose minimal medium unless it is supplemented with acetate and succinate to bypass the requirement for PDH and α-ketoglutarate dehydrogenase (5, 13).
Bypassing LipB.
The de novo biosynthetic pathway in E. coli requires lipA and lipB (Fig. 2A). Genetic studies identified the product of the lipA gene as responsible for inserting the two sulfur atoms into the octanoate precursor (13). LipA is an S-adenosylmethionine-dependent enzyme that inserts sulfur at carbons 6 and 8 of octanoate using a radical mechanism (9). A key discovery was that LipA introduces the sulfur atoms into octanoate bound to the E2 subunit of PDH and not into free octanoic acid or octanoyl-ACP (15). Thus, the de novo biosynthetic pathway begins by the diversion of octanoyl-ACP from the type II fatty acid synthesis cycle by LipB, an acyltransferase that transfers the octanoyl moiety from ACP to the lysine acceptor in the E2 domain via an acyl enzyme intermediate (14). The most straightforward route to lipoyl enzymes is the salvage pathway, which involves uptake of environmental lipoic acid followed by its activation and transfer to the E2 domains (Fig. 2B). This two-step reaction is performed by LplA. The first step is the ATP-dependent formation of an LplA-bound lipoyl-AMP intermediate, followed by transfer of lipoate to the E2 domain by its acyltransferase activity.
Whereas LplA has a high affinity for lipoate, LplA also weakly activates several lipoate analogs and octanoate for transfer to the E2 apoproteins (4, 9). This lack of substrate specificity allows LipB null mutants to grow in the presence of either a lipoate or an octanoate supplement (15). The strategy employed by Hermes and Cronan was to select for variants of a lipB null strain that were able to grow without an exogenous source of lipoate or octanoate (6). Two such strains were isolated. They quickly determined that the mutations were in lplA and that the modified genes encoded either the LplA(V19L) or LplA(S221P) mutant protein. Strains with the suppressor mutations contain detectable quantities of lipoyl domains, and the growth of the suppressor mutants still depends on a functional lipA gene. The discovery that the affinity of LplA for octanoate was increased by these point mutations suggested that the LplA mutant proteins activated an intracellular source of octanoate to bypass LipB function (Fig. 2C).
LplA mutants and protein structure.
The LplA proteins consist of two domains, but they are not always organized in the same manner. E. coli LplA has a large amino-terminal domain and a smaller carboxy-terminal domain (3). In Streptomyces coelicolor, the carboxy-terminal domain of E. coli LplA is predicted at the amino terminus. The crystal structures of LplA from the archaeon Thermoplasma acidophilum lack the characteristic carboxy-terminal domain (7, 8). This conundrum was recently solved by the demonstration that the carboxy-terminal domain of E. coli LplA belongs to a separate protein, called LplB, in T. acidophilum (1). Biochemical experiments show that the LplB domain is required for lipoyl-adenylate formation, although lipoyl-AMP is bound to the LplA subunit and can be transferred to E2 domains in the absence of LplB.
The LplA(V19L) mutant was isolated four times in suppressor strains created by random mutagenesis using error-prone PCR, indicating its importance in influencing substrate specificity. An earlier selection for LplA mutants resistant to seleno-lipoate, an analog with sulfur atoms replaced with selenium, yielded an LplA(G76S) protein (12). E. coli Gly76 is located on a loop that is thought to interact with the dithiolane ring of lipoic acid in the LplA structures from T. acidophilum (7, 8). It is easy to envision how the introduction of the serine side chain may sterically hinder the binding of the bulky seleno-lipoate analog (8). However, the interpretation of how LplA(V19L) has increased affinity for octanoate compared to that of the wild-type protein is not as obvious. Although Val19 is close to the lipoate binding site in the T. acidophilum structure, the side chain points away from the lipoyl group, and Val19 is not a conserved residue in LplA proteins. The mutation is therefore likely to affect the overall dimensions of the active site by altering the packing of the structural elements. Consistent with this idea, the overall catalytic activity of LplA(V19L) is lower than that of the wild type. While the genetic approach clearly identifies the suppressor mutations as being involved in substrate specificity, additional crystal structures that trap LplA in the different conformations will be required to determine how these amino acid modifications alter the LplA substrate site.
Enigmatic origin and function of intracellular fatty acids.
A surprising outcome from this work is the detection of medium-chain free fatty acids in E. coli. The isolation of lplA suppressor mutants with increased affinities for octanoate clearly pointed to the presence of an intracellular pool of this fatty acid. There was a hint that such a pool may exist from earlier work showing that LplA overexpression suppressed the growth defect of lipB null mutants (10), but free fatty acids, especially of the medium-chain variety, were not thought to exist in vivo. Hermes and Cronan used contemporary analytical techniques to trap and quantify these volatile fatty acids to prove their presence in vivo. It seems most likely that the fatty acids are derived from fatty acid synthesis, presumably by a thioesterase activity on the acyl-ACP intermediates (Fig. 2C). Hermes and Cronan tested the usual thioesterase suspects, TesA and TesB, but knocking out the genes encoding these thioesterases did not abrogate the ability of LplA suppressor strains to grow in the absence of a supplement. Perhaps one of the thioesterase-like genes in E. coli without an assigned function is responsible for the generation of the intracellular free fatty acids. It is also possible that their occurrence reflects an inherent inefficiency in intermediary metabolism due to nonspecific acyl-ACP hydrolysis by multiple thioesterases or low-level hydrolysis of the β-ketoacyl-ACP synthase acyl-enzyme intermediates by water. This idea suggests that all chain lengths of free fatty acids may be present at some level in vivo. Hermes and Cronan show that multiple medium-chain fatty acids are present in the cell, indicating a process that is not chain length specific. More data are needed to sort out these possibilities, but while it is clear that free octanoate is not normally an intermediate in lipoic acid synthesis, the larger question of whether or not these fatty acids are produced for another unknown cellular function(s) remains open.
Acknowledgments
Work in my laboratory is supported by National Institutes of Health grants GM 34496 and Cancer Center (CORE) support grant CA 21765 and the American Lebanese Syrian Associated Charities.
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
Footnotes
Published ahead of print on 4 September 2009.
REFERENCES
- 1.Christensen, Q. H., and J. E. Cronan. 2009. The Thermoplasma acidophilum LplA-LplB complex defines a new class of bipartite lipoate-protein ligases. J. Biol. Chem. 284:21317-21326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cronan, J. E., X. Zhao, and Y. Jiang. 2005. Function, attachment and synthesis of lipoic acid in Escherichia coli. Adv. Microb. Physiol. 50:103-146. [DOI] [PubMed] [Google Scholar]
- 3.Fujiwara, K., S. Toma, K. Okamura-Ikeda, Y. Motokawa, A. Nakagawa, and H. Taniguchi. 2005. Crystal structure of lipoate-protein ligase A from Escherichia coli. Determination of the lipoic acid-binding site. J. Biol. Chem. 280:33645-33651. [DOI] [PubMed] [Google Scholar]
- 4.Green, D. W., T. W. Morris, J. Green, J. E. Cronan, Jr., and J. R. Guest. 1995. Purification and properties of the lipoate protein ligase of Escherichia coli. Biochem. J. 309:853-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herbert, A. A., and J. R. Guest. 1968. Biochemical and genetic studies with lysine+methionine mutants of Escherichia coli: lipoic acid and α-ketoglutarate dehydrogenase-less mutants. J. Gen. Microbiol. 53:363-381. [DOI] [PubMed] [Google Scholar]
- 6.Hermes, F. A., and J. E. Cronan. 2009. Scavenging of cytosolic octanoic acid by mutant LplA lipoate ligases allows growth of Escherichia coli strains lacking the LipB octanoyltransferase of lipoic acid synthesis. J. Bacteriol. 191:6796-6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim, D. J., K. H. Kim, H. H. Lee, S. J. Lee, J. Y. Ha, H. J. Yoon, and S. W. Suh. 2005. Crystal structure of lipoate-protein ligase A bound with the activated intermediate: insights into interaction with lipoyl domains. J. Biol. Chem. 280:38081-38089. [DOI] [PubMed] [Google Scholar]
- 8.McManus, E., B. F. Luisi, and R. N. Perham. 2006. Structure of a putative lipoate protein ligase from Thermoplasma acidophilum and the mechanism of target selection for post-translational modification. J. Mol. Biol. 356:625-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morris, T. W., K. E. Reed, and J. E. Cronan, Jr. 1994. Identification of the gene encoding lipoate-protein ligase A of Escherichia coli. Molecular cloning and characterization of the lplA gene and gene product. J. Biol. Chem. 269:16091-16100. [PubMed] [Google Scholar]
- 10.Morris, T. W., K. E. Reed, and J. E. Cronan, Jr. 1995. Lipoic acid metabolism in Escherichia coli: the lplA and lipB genes define redundant pathways for ligation of lipoyl groups to apoprotein. J. Bacteriol. 177:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perham, R. N. 2000. Swinging arms and swinging domains in multifunctional enzymes: catalytic machines for multistep reactions. Annu. Rev. Biochem. 69:961-1004. [DOI] [PubMed] [Google Scholar]
- 12.Reed, K. E., T. W. Morris, and J. E. Cronan, Jr. 1994. Mutants of Escherichia coli K-12 that are resistant to a selenium analog of lipoic acid identify unknown genes in lipoate metabolism. Proc. Natl. Acad. Sci. USA 91:3720-3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vanden Boom, T. J., K. E. Reed, and J. E. Cronan, Jr. 1991. Lipoic acid metabolism in Escherichia coli: isolation of null mutants defective in lipoic acid biosynthesis, molecular cloning and characterization of the E. coli lip locus, and identification of the lipoylated protein of the glycine cleavage system. J. Bacteriol. 173:6411-6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao, X., J. R. Miller, and J. E. Cronan. 2005. The reaction of LipB, the octanoyl-[acyl carrier protein]:protein N-octanoyltransferase of lipoic acid synthesis, proceeds through an acyl-enzyme intermediate. Biochemistry 44:16737-16746. [DOI] [PubMed] [Google Scholar]
- 15.Zhao, X., J. R. Miller, Y. Jiang, M. A. Marletta, and J. E. Cronan. 2003. Assembly of the covalent linkage between lipoic acid and its cognate enzymes. Chem. Biol. 10:1293-1302. [DOI] [PubMed] [Google Scholar]