Abstract
The bacterium Helicobacter pylori is remarkable for its ability to persist in the human stomach for decades without provoking sterilizing immunity. Since repetitive DNA can facilitate adaptive genomic flexibility via increased recombination, insertion, and deletion, we searched the genomes of two H. pylori strains for nucleotide repeats. We discovered a family of genes with extensive repetitive DNA that we have termed the H. pylori RD gene family. Each gene of this family is composed of a conserved 3′ region, a variable mid-region encoding 7 and 11 amino acid repeats, and a 5′ region containing one of two possible alleles. Analysis of five complete genome sequences and PCR genotyping of 42 H. pylori strains revealed extensive variation between strains in the number, location, and arrangement of RD genes. Furthermore, examination of multiple strains isolated from a single subject's stomach revealed intrahost variation in repeat number and composition. Despite prior evidence that the protein products of this gene family are expressed at the bacterial cell surface, enzyme-linked immunosorbent assay and immunoblot studies revealed no consistent seroreactivity to a recombinant RD protein by H. pylori-positive hosts. The pattern of repeats uncovered in the RD gene family appears to reflect slipped-strand mispairing or domain duplication, allowing for redundancy and subsequent diversity in genotype and phenotype. This novel family of hypervariable genes with conserved, repetitive, and allelic domains may represent an important locus for understanding H. pylori persistence in its natural host.
Helicobacter pylori, a gram-negative bacterium, is remarkable for its ability to persist in the human stomach for decades. Colonization with H. pylori increases risk for peptic ulcer disease and gastric adenocarcinoma (53, 70) and elicits a vigorous immune response (15). The persistence of H. pylori occurs in a niche in the human body previously considered inhospitable to microbial colonization: the acidic stomach replete with proteolytic enzymes.
H. pylori strains exhibit substantial genetic diversity, including extensive variation in the presence, arrangement, order, and identity of genes (2, 4-7, 25, 51, 74). Furthermore, analyses of multiple single-colony H. pylori isolates from separate stomach biopsy specimens of individual patients have demonstrated diversity, both within hosts (27, 65), and over time (36). The mechanisms that generate H. pylori genetic diversity may be among the factors that enable persistence in this environment (3, 28).
While the natural ability of H. pylori for transformation and recombination may explain some of the intra- and interhost genetic variation observed in this bacterium (43), point mutations and interspecies recombination alone are not sufficient for explaining the extent of the variation in H. pylori (14, 32). The initial genomic sequencing of H. pylori strains 26695 and J99 (6, 72) revealed large amounts of repetitive DNA (1, 59). DNA repeats in bacteria are associated with mechanisms of plasticity, such as phase variation (49, 67); slipped-strand mispairing (41, 46); and increased rates of recombination, deletion, and insertion (17, 60, 62). Because many of the recombination repair and mismatch repair mechanisms common in bacteria are absent or modified in H. pylori (28-30, 56, 76), this organism may be particularly susceptible to the diversifying effects of repetitive DNA. In fact, loci in the H. pylori genome containing repetitive DNA have been shown to exhibit extensive inter- and intrahost variation (9, 10, 28, 37).
We hypothesized that identification of repetitive DNA hotspots in H. pylori would allow the recognition of genes whose variation could aid in persistence. To examine this hypothesis, we conducted in silico analyses to identify open reading frames (ORFs) enriched for DNA repeats and then used a combination of sequence analyses and immunoassays to examine the patterns associated with the specific repetitive DNA observed. Our approach led to the realization that a previously identified H. pylori-specific gene family (19, 52) exhibits extensive genetic variation at multiple levels.
MATERIALS AND METHODS
Bacterial strains.
Genomic sequences of Helicobacter pylori strains 26695 (72), J99 (6), and HPAG1 (51) were retrieved from GenBank (12), and specific sequences from the H. pylori G27 genome were provided ahead of publication (11). In addition, 43 H. pylori strains from Asia, Europe, North America, and South America from patients with differing clinical diagnoses were studied (see Table S1 in the supplemental material). These strains, stored in the New York University Helicobacter/Campylobacter strain reference collection at −70°C, were cultured at 37°C in a 5% CO2 atmosphere on Trypticase soy agar plates with 5% sheep blood. For one of these strains, HP1 (26), nucleotide sequencing of specific genes of interest was conducted. In addition, multiple H. pylori isolates were obtained from patients in Ladakh, India, during endoscopy (63).
Computational analyses.
Identification of forward, perfect nucleotide repeats of >24 base pairs in strains 26695 and J99 was done using the computer program REPuter (39). We chose a minimum length of 25 because the probability of finding repeats of this length by chance alone in the H. pylori genome is <0.001 (59). To assess homology, Clustal X version 2.0 (40) was used to align DNA sequences and Swaap 1.0.3 (57) was used to calculate pairwise nucleotide and amino acid identities as well as synonymous- and nonsynonymous-substitution rates (Ks and Ka, respectively). Kyte-Doolittle hydropathy plots were created with FASTA at the University of Virginia (http://fasta.bioch.virginia.edu/fasta_www2/fasta_www.cgi?rm=misc1). Amino acid sequences were examined for protein sequence motifs and predicted coiled-coil regions by using the Motifs and Coilscan programs, respectively, in SeqWeb version 3.1. For the Coilscan analysis, a 28-amino-acid sliding window was used to determine the coiled-coil probability for each residue. Phylogenetic analyses were conducted with PAUP* version 4.0b10 (71), using the distance criterion. We employed a neighbor-joining technique and estimated support for individual branches by conducting 1,000 bootstrap replicates (21).
To achieve a working classification, repeat units (all 21 or 33 bp) were identified and aligned using the Clustal algorithm within MEGA version 3.1 (38). Since the alignments were very short, distances between fragments were determined by absolute number of differences, and a tree based on the unweighted-pair group method using average linkages was constructed using MEGA version 3.1 (38) with 1,000 repetitions to determine bootstrap values.
To evaluate the possibility of recombination events, sequences of the gene regions of interest were aligned and then evaluated with the Sawyer run test, implemented in GENECONV (68, 69).
RD genotyping.
After 72 h of bacterial growth on Trypticase soy agar plates with 5% sheep blood, genomic DNA was prepared from each studied strain. PCRs for determining RD genotypes were conducted with 1× buffer, 0.20 mM deoxynucleoside triphosphates, 0.40 μM of each primer, 0.5 units of Taq polymerase, and 100 ng DNA in a 50-μl total volume. The thermal cycler program used an initial denaturation step at 94°C for 3 min; 30 cycles at 94°C for 30 s, 60°C for 30 s, and 72°C for 4 min; and a final extension at 72°C for 10 min. Product length was determined by agarose gel electrophoresis. The number of RD genes at each locus was determined with amplifications using primers RDL1-5 and RDL1-3, located in the conserved genes flanking RD locus 1, and primers RDL2-5 and RDL2-3, located in the conserved genes flanking RD locus 2 (see Table S2 in the supplemental material). The allelic identities of RD genes of the 5′ allelic region (FAR) were determined by sequencing or by amplification with primers placed in FAR1 (FAR1f and FAR1r) or FAR2 (FAR2f and FAR2r) (see Fig. S1 in the supplemental material).
For the strains isolated from patients in Ladakh, India, DNA fingerprinting was conducted by random amplified polymorphic DNA-PCR with 10-nucleotide primers 1254, 1281, and 1290, as described previously (5, 78).
Expression of JHP_0110.
Primers were designed to amplify the full-length JHP_0110 gene from H. pylori strain J99, with addition of a 5′ XhoI restriction site and a 3′ BamHI restriction site (see Table S2 in the supplemental material). PCRs were prepared as described above, but the thermal cycler program had an initial denaturation step at 94°C for 3 min; 30 cycles at 94°C for 1 min, 58°C for 1 min, and 72°C for 1 min; and a final extension at 72°C for 10 min.
PCR products treated with a QIAquick PCR purification kit (Qiagen, Valencia, CA) and pET-15b vector were digested with both XhoI and BamHI, and molecular cloning was done according to the manufacturer's guidelines (EMD Biosciences, Darmstadt, Germany). QIAprep spin miniprep kits (Qiagen, Inc., Valencia, CA) were used to isolate plasmid from XL1-blue Escherichia coli cells. The pet15b-JHP_0110 construct was transformed into BL21(DE3) E. coli cells for expression, and the recombinant protein was isolated from inclusion bodies, in accordance with the Novagen Bugbuster protocol. The Novagen His bind kit (EMD Biosciences, Darmstadt, Germany) was used to purify the recombinant protein under denaturing conditions (6 M urea).
Serologic assays.
Sera from a previously described cohort of H. pylori-negative children (54) were used as negative controls. We studied the sera from 247 adults undergoing endoscopy at the VA New York Harbor Healthcare System. Patients were considered H. pylori positive if they showed positive results for at least two of the following assays: histology, culture, serology, and rapid urease testing (24). In brief, enzyme-linked immunosorbent assays (ELISAs) were conducted by fixing 10 ng of recombinant full-length JHP_0110 protein to each well, using carbonate buffer (pH 9.6). After being blocked, samples were added to wells in duplicate. The secondary antibody, goat anti-human immunoglobulin G (IgG)-horseradish peroxidase conjugate, was diluted 1:4,000 in phosphate-buffered saline containing 0.05% Tween 20, 0.02% thimerosal, 0.1% gamma globulin, and 1.0% albumin and incubated in wells for 1 h at 37°C. Developer containing 45% Na2HPO4, 55% citric acid, 0.16% H2O2, and 10 mg of ABTS [2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid)] was added and optical density (OD) measured at 405 nm. For immunoblots, recombinant full-length JHP_0110 protein in 6 M urea was electrophoresed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane. Human sera were diluted 1:3,000 and mouse anti-His diluted 1:2,000. Goat anti-human IgG-alkaline phosphatase conjugate, diluted 1:1,000, and BCIP (5-bromo-4-chloro-3-indolylphosphate)-nitroblue tetrazolium phosphate substrate were used to visualize the conjugated protein.
RESULTS
Relatively few H. pylori ORFs contain the majority of direct repeats.
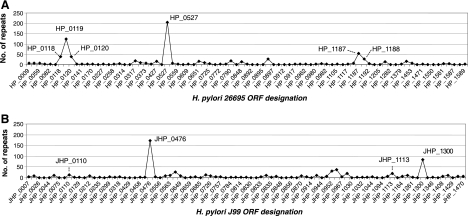
An analysis of direct identical repeats of >24 bp by use of the REPuter program revealed 554 repeats in the H. pylori 26695 genome and 482 repeats in the H. pylori J99 genome (Fig. 1). As expected (10), cagY was the ORF in both the 26695 and the J99 genomes (HP_0527 and JHP_0476, respectively) with the most repeats. Only six other ORFs in strain 26695 contained >20 direct identical repeats. Five of these ORFs in strain 26695 (HP_0118, HP_0119, HP_0120, HP_1187, and HP_1188) and two ORFs in strain J99 (JHP_0110 and JHP_1113) all belonged to a single gene family (DUF874) according to Pfam (22). Because of their repetitive DNA sequences, we propose calling this group of ORFs the H. pylori “RD gene family.”
FIG. 1.
DNA repeats per ORF in the genomes of H. pylori strains J99 and 26695. Totals of 554 and 482 forward repeats of >24 bp were observed within ORFs in strains 26695 and J99, respectively. In both genomes, the most repeats were observed in the ORFs encoding the CagY protein (HP_0527 and JHP_0476 [61]). The second-largest peak in the J99 genome, JHP_1300, encodes a putative protein of unknown function. Other ORFs with large numbers of repeats (HP_0118, HP_0119, HP_0120, HP_1187, HP_1188, JHP_0110, and JHP_1113) shared substantial homology with one another and now have been termed H. pylori “RD genes.”
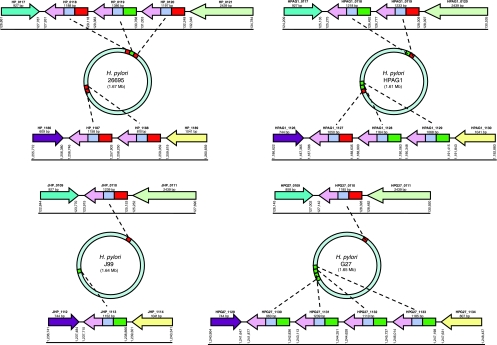
We termed the location of the three adjacent ORFs in 26695 (HP_0118, HP_0119, and HP_0120) RD locus 1 and the location of the two other adjacent ORFs (HP_1187 and HP_1188) RD locus 2 (Fig. 2). In strain J99, RD locus 1 contains JHP_0110, and RD locus 2 contains JHP_1113. Strain HPAG1 has two genes at RD locus 1 and three at RD locus 2 (five in total), whereas G27 has one gene at RD locus 1 and four genes at RD locus 2 (also five in total) (Fig. 2). In the four studied H. pylori strains with completely sequenced genomes, RD locus 1 was always flanked by a hypothetical protein (HP_0117) and phosphoenolpyruvate synthase (HP_0121), and RD locus 2 was always flanked by carbonic anhydrase (HP_1186) and aspartate-semialdehyde dehydrogenase (HP_1189) (Fig. 2). For another H. pylori strain, HPI, studied using flanking and internal primers, two RD genes (one at each locus, resembling J99) were identified and sequences analyzed. In total, the 19 RD genes identified in these five strains were the substrate for further studies.
FIG. 2.
Schematic of RD genes in H. pylori strains 26695, J99, HPAG1, and G27. The conserved 3′ region found in all RD genes is indicated in lilac, the two types of FARs are indicated in red (FAR1) and green (FAR2), and the variable mid-region is indicated in blue. Flanking genes with homology to one another are indicated by color: turquoise (HP_0117 and homologs), light green (HP_0121 and homologs), purple (HP_1186 and homologs), and yellow (HP_1189 and homologs).
RD genes share a common structure.
All 19 RD genes examined had a FAR, a conserved 3′ region, and a variable mid-region (Fig. 2). The FAR of each RD gene contained one of two alleles, designated FAR1 and FAR2, which share 32.6% ± 1.1% pairwise nucleotide identity (Table 1). In contrast, the nucleotide sequences of the FARs in HP_0118, HP_0120, HP_1187, HP_1188, JHP_0110, HPAG1_0119, HPAG1_1127, G27_0110, and HP1_L1 were 93.9% ± 1.6% identical (mean ± SD) (Table 1); we termed this allele FAR1 (amino acid positions 1 to 122 in JHP_0110) (Fig. 2). Similarly, the nucleotide sequences of the FARs in HP_0119, JHP_1113, HPAG1_0118, HPAG1_1128, HPAG1_1129, G27_1130, G27_1131, G27_1132, G27_1133, and HP1_L2 were 97.2% ± 1.0% identical (Table 1); we termed this allele FAR2 (amino acid positions 1 to 110 in JHP_1113) (Fig. 2). FAR2 sequences shared significantly more (P < 0.001) identity than FAR1 sequences at both the nucleotide and amino acid levels (Table 1), but FARs from the same strain were no more identical at the nucleotide or amino acid levels than FARs from different strains (Table 1).
TABLE 1.
Analysis of identity and substitutions for 19 H. pylori RD genes in five studied H. pylori strainsa
| Comparison | No. of comparisonsc | FAR sequencesb |
3′ region sequencesb |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| % Identity |
Ka | Ks | Ka/Ks | % Identity |
Ka | Ks | Ka/Ks | ||||
| Nucleotide | Amino acid | Nucleotide | Amino acid | ||||||||
| FAR1 genes vs FAR1 genes | 36 | 93.9 ± 1.6* | 91.2 ± 2.3* | 0.04 ± 0.01* | 0.14 ± 0.06* | 0.28 ± 0.13 | 94.7 ± 1.2 | 90.9 ± 2.5 | 0.05 ± 0.01 | 0.10 ± 0.03 | 0.51 ± 0.24 |
| FAR2 genes vs FAR2 genes | 45 | 97.2 ± 1.0 | 95.8 ± 1.9 | 0.02 ± 0.01 | 0.08 ± 0.04 | 0.27 ± 0.19 | 94.6 ± 1.9 | 90.9 ± 3.0 | 0.05 ± 0.01 | 0.11 ± 0.05 | 0.44 ± 0.18 |
| FAR1 genes vs FAR2 genes | 90 | 32.6 ± 1.1 | 10.2 ± 1.5 | 1.17 ± 0.54 | 0.88 ± 0.14 | 1.30 ± 0.30 | 94.6 ± 1.3 | 90.7 ± 2.3 | 0.05 ± 0.01 | 0.11 ± 0.04 | 0.47 ± 0.20 |
| FAR1 genes (intrastrain) | 7 | 95.7 ± 2.2 | 93.8 ± 3.1 | 0.03 ± 0.02 | 0.07 ± 0.06* | 0.18 ± 0.17 | 95.3 ± 1.1 | 92.0 ± 2.1 | 0.04 ± 0.01 | 0.08 ± 0.02 | 0.54 ± 0.19 |
| FAR1 genes (interstrain) | 29 | 93.5 ± 1.1 | 90.5 ± 1.6 | 0.04 ± 0.01 | 0.16 ± 0.04 | 0.30 ± 0.10 | 94.5 ± 1.2 | 90.6 ± 2.6 | 0.05 ± 0.01 | 0.10 ± 0.03 | 0.50 ± 0.26 |
| FAR2 genes (intrastrain) | 9 | 97.6 ± 1.8 | 96.4 ± 3.0 | 0.02 ± 0.01 | 0.06 ± 0.04 | 0.20 ± 0.13 | 96.9 ± 2.0* | 94.9 ± 3.0* | 0.03 ± 0.01* | 0.07 ± 0.05 | 0.46 ± 0.35 |
| FAR2 genes (interstrain) | 36 | 97.0 ± 0.7 | 95.7 ± 1.5 | 0.02 ± 0.01 | 0.08 ± 0.04 | 0.29 ± 0.20 | 94.0 ± 1.3 | 89.9 ± 2.0 | 0.05 ± 0.01 | 0.12 ± 0.04 | 0.43 ± 0.11 |
| All comparisons | 171 | 62.5 ± 31.6 | 49.8 ± 41.9 | 0.63 ± 0.69 | 0.51 ± 0.40 | 0.81 ± 0.57 | 94.6 ± 1.5 | 90.8 ± 2.1 | 0.05 ± 0.01 | 0.11 ± 0.04 | 0.47 ± 0.20 |
Genome-sequenced strains 26695, J99, HPAG1, and G27 and studied strain HP1.
Values shown are means ± standard deviations of results from all pairwise comparisons. * signifies P values of <0.01 for comparison of the value with the value directly below.
t tests were used to test for significant differences between results for FAR1 gene comparisons and FAR2 gene comparisons as well as between results for intra- and interstrain comparisons.
There was substantial homology across all strains in the 3′ region (amino acid positions 228 to 412 in JHP_0110 and 199 to 383 in JHP_1113). The 19 RD genes in strains 26695, J99, HPAG1, G27, and HP1, had 3′ regions that were 94.6% ± 1.5% identical at the nucleotide level and 90.8% ± 2.1% identical at the amino acid level (Table 1). At the nucleotide level, the identities between the 3′ regions of the genes with FAR1 (94.7% ± 1.2%) did not differ significantly (P = 0.73) from the identities between the 3′ regions of the genes with FAR2 (94.6% ± 1.9%) (Table 1). Similarly, at the amino acid level, the identities between 3′ regions of the genes with FAR1 (90.9% ± 2.5%) did not differ significantly (P = 0.95) from the identities between the 3′ regions of the genes with FAR2 (90.9% ± 3.0%) (Table 1). The nucleotide and amino acid identities of the 3′ region for intrastrain comparisons were significantly greater than those for interstrain comparisons for genes containing FAR2 and trended in that direction for FAR1.
Evolution of the 3′ regions and FARs.
The 19 3′ region sequences studied showed a low Ks (0.11 ± 0.04), similar to that for H. pylori housekeeping genes (2), regardless of the FAR allele of its gene and regardless of whether the RD genes were in the same strain. Ka was also quite low, regardless of type or origin of the 3′ region. Overall, on the basis of 171 pairwise comparisons, the 3′ regions of the 19 RD genes in the five studied strains (26695, J99, HPAG1, G27, and HP1) had a Ka/Ks ratio of 0.47 ± 0.20 (mean ± SD) (Table 1). The Ka/Ks ratios were found to be 0.51 ± 0.24 when the 3′ regions of RD genes with FAR1 were compared, 0.44 ± 0.18 when the 3′ regions of RD genes with FAR2 were compared, and 0.47 ± 0.20 when the 3′ regions of RD genes with different FARs were compared (Table 1). While Ka was significantly higher (P = 0.001) for the 9 FAR1 sequences (0.04 ± 0.01) than for the 10 FAR2 sequences (0.02 ± 0.01), the Ka/Ks ratios for FAR1 sequences and FAR2 sequences were not significantly different (P = 0.86) (Table 1). That the Ka/Ks ratio for all 3′ region comparisons (0.47 ± 0.20) was significantly higher (P < 0.001) than the Ka/Ks ratios for FAR1 (0.28 ± 0.13) and FAR2 (0.27 ± 0.19) comparisons is consistent with the idea that there is greater selective pressure for variation on the 3′ region than for variation on individual FARs. The intra-allele substitution rates for the FAR sequences and those for the 3′ regions were similarly low, but the 90 FAR1/FAR2 comparisons showed high substitution rates and evidence for diversifying selection (Ka/Ks = 1.30 ± 0.30) (Table 1).
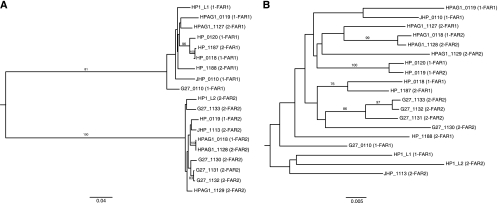
Separate phylogenetic analyses were conducted using nucleotide sequence data from the FARs and the 3′ regions of the 19 RD genes in the five studied strains (Fig. 3). The tree constructed using FAR sequences (Fig. 3A) contains two strongly supported branches, with one containing all 9 FAR1 sequences and one containing all 10 FAR2 sequences. Within these two groups, RD genes were not monophyletic on the basis of strain or RD locus. In the tree constructed from 3′ region sequences (Fig. 3B), RD genes were not monophyletic on the basis of FAR allele, RD locus, or strain. All of the strongly supported branches (bootstrap values of >70) were for sequences within the same strain, consistent with concerted evolution facilitated by gene conversion (57). All branches on both the FAR and the 3′ region nucleotide trees with bootstrap support values of >70 also appeared on neighbor-joining trees inferred from amino acid data (not shown). Thus, the evolution of the two strongly conserved domains of the RD genes, the FAR and 3′ region, show evidence of substantially different selection.
FIG. 3.
Phylograms of the 19 RD genes in H. pylori strains HP1, 26695, J99, HPAG1, and G27, inferred using nucleotide data from the FAR (A) and the 3′ region (B). Trees were inferred by neighbor-joining analysis and rooted at the midpoint. Bootstrap values of >70 are indicated at branch points. The RD locus of each gene is indicated by the first number in parentheses, and the FAR type is indicated by the second number.
Variation in RD genotype among H. pylori strains.
Using a PCR-based technique to determine the contents of each RD locus (see Fig. S1 in the supplemental material), we found extensive diversity in the number and arrangement of RD genes in 47 H. pylori strains from various parts of the world (see Table S1 in the supplemental material). Nevertheless, three conserved patterns were identified: (i) for each strain studied, each RD locus contained at least one RD gene; (ii) there were no empty sites; and (iii) no strain had more than five RD genes combined between the two loci. The numbers of RD genes per strain in the eight Cag-negative strains (2.38 ± 0.52) and in the 30 Cag-positive strains (2.57 ± 0.97) were nearly identical (P = 0.46). The most common genotype, present in 34.0% of strains, was a FAR1 gene at locus 1 and a FAR2 gene at locus 2 (see Fig. S2 in the supplemental material). Of the strains studied, 68.1% had two RD genes, 23.4% had three RD genes, 2.1% had four RD genes, and 6.4% had five RD genes. In total, of the 116 RD genes present in the 47 strains, 59 (50.9%) were at locus 1 and 57 (49.1%) were at locus 2. Of the genes at locus 1, 67.8% are FAR1, whereas at locus 2, 36.8% are FAR1, an allelic distribution unlikely to be due to chance alone (χ2 = 5.7; P = 0.017). Of the 17 strains with single FAR1 and FAR2 genes, 16 (94%) had FAR1 at locus 1 and FAR2 at locus 2 (χ2 = 13.2; P = 0.0003). Thus, although RD gene distribution varied greatly among strains, several strongly nonrandom organizational principles were observed.
Recombination within the FAR and 3′ region.
When the Sawyer run algorithms were used to detect putative recombination events in the 19 FAR segments, no statistically significant indications of recombination were found within the FAR1 or the FAR2 or between them. However, the same analysis of the 3′ region revealed 6 putative recombinant fragments with global statistical support for recombination and 43 putative inner fragments with pairwise support (see Table S3 in the supplemental material). Of these 49 possible recombination events, 26 were identified between differing FAR types (FAR1 or FAR2) and 27 were identified between different strains of H. pylori (see Table S4 in the supplemental material). These data support the phylogenetic studies suggesting gene conversion.
Variation in composition and order of repeats in mid- regions.
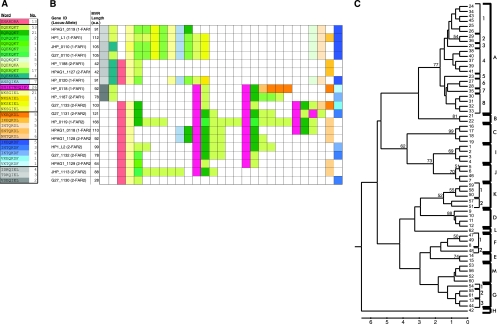
The mid-region shows extensive variation. However, several conserved principles still emerge. Among the RD genes in the five studied strains, the mid-regions ranged in size from 28 to 166 encoded amino acids and were composed of imperfect 7- and 11-amino-acid repeats (“words”). For illustration and analysis, each word was assigned a color, with similar words (with three or fewer amino acid mismatches) designated shades of the same color (Fig. 4A), and the mid-regions of the five strains were manually aligned (Fig. 4B).
FIG. 4.
Schematic of the 229 repeat “words” in the mid-regions of the 19 RD genes in H. pylori strains 26695, J99, HPAG1, G27, and HP1. (A) Color coding of conserved words. Words that appear in the mid-regions of these 19 RD genes, along with their assigned color code and the number of times each word appears. (B) Mid-region organization by word type and number. Each color-coded box represents a 7- or 11-amino-acid-repeat word. Boxes have been manually aligned to demonstrate patterns shared among mid-regions. (C) Phylogeny of 62 individual words. Each unique 21- or 33-nucleotide fragment received an identifier that included a one- or two-digit number followed by a group designation (indicated by a capital letter) and a subgroup designation (indicated by a single-digit number).
Of the 229 words identified, there were 62 unique nucleotide sequences, with a nonrandom distribution of their prevalence (Fig. 4A). In contrast to the FAR domain, no strong phylogenetic signal was seen in the mid-region (Fig. 4C). To compensate for poor bootstrap values in the tree clustering, each fragment was assigned to a group and in some cases to a subgroup (Fig. 4C). The subgroups are ≤4 bp different from one another, predominantly ≤3 bp, consistent with clusters with poor bootstrap values. Several relatively well-differentiated clusters (with bootstrap values of ≥50) were identified. Most members of a single group are ≤6 bp different from each other and are variably differentiated from other groups. A pattern of great complexity emerged (see Tables S4 and S5 in the supplemental material); the large number of near-repeats instead of exact repeats was unexpected. Group A sequences were the most prevalent, and FAR2 sequences were more likely to have repeats from group A than were the FAR1 sequences (P = 0.017 by Fisher's exact test). There were no repeats within a single gene for fragments in eight groups (B, C, E, F, H, K, L, and M). No RD genes containing FAR1 had group D repeats except sequences HP_0118 and HP_1187, and the distributions of alleles in group A clearly differed in FAR1 and FAR2 sequences. The mid-regions of HP_0118 and HP_1187 resembled FAR2 mid-regions and may represent a recombinant event. Thus, although the mid-regions have many conserved features in aggregate, the mid-region is a locus of great genetic diversity.
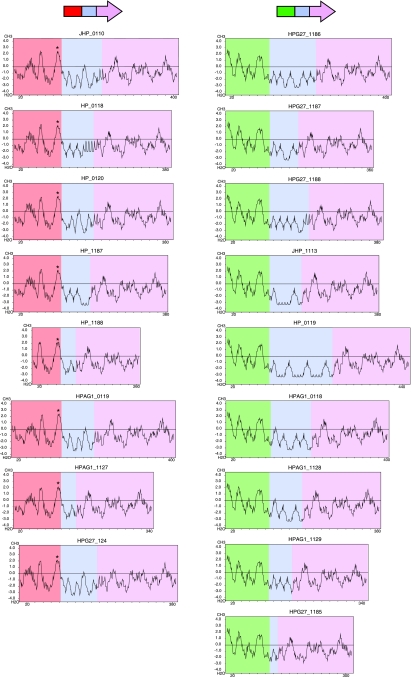
The heptad repeats of the mid-region display characteristics consistent with a coiled-coil section of protein (16, 47). Kyte-Doolittle hydropathy analyses demonstrate that the mid-regions of the RD genes are mostly hydrophilic (Fig. 5), which can be explained by the high frequency of amino acids with polar side chains including glutamic acid (E), glutamine (Q), asparagine (N), and lysine (K) in the mid-region words (Fig. 4A). Coilscan analyses of the 19 RD genes in the five studied strains predicted that 91.0% of the 1,693 amino acids in mid-regions are part of a coiled-coil structure, compared to 7.5% of the 2,154 amino acids in FARs and 4.5% of the 3,465 amino acids in 3′ regions (P < 0.001 for both comparisons). Thus, despite the primary sequence diversity, there is strong conservation of secondary structure. Consistent with the FAR allelic differences, putative prokaryotic membrane lipoprotein lipid attachment sites were identified in all FAR1 sequences in strains 26696, J99, HPAG1, and G27 but not in any FAR2 sequences (Fig. 5).
FIG. 5.
Kyte-Doolittle hydropathy plots of RD genes in H. pylori strains J99, 26695, HPAG1, and G27. 3′ regions are colored lilac, mid-regions are colored blue, FAR1 areas are colored red, and FAR2 areas are colored green. * indicates a prokaryotic membrane lipoprotein lipid attachment site, as identified by the Motifs program in SeqWeb.
Variation in mid-regions of human H. pylori isolates.
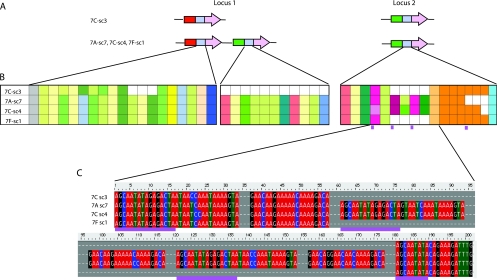
To begin to examine intrahost variation, RD genotyping and mid-region sequencing was performed for four H. pylori stomach isolates (one from the antrum, two from the corpus, and one from the fundus) obtained from a patient from Ladakh, India. Many of the words in the mid-regions of the five studied strains (Fig. 4A) also were present in the mid-regions of the Ladakh isolates (Fig. 6). At locus 1, single colony 3 from the corpus had only one RD gene, whereas the three other isolates had two (Fig. 6A). The three strains (antrum-sc7, corpus-sc4, and fundus-sc1) that had two RD genes at locus 1 did not have identical mid-regions at locus 2 (Fig. 6B). Thus, all four isolates had differing RD genotypes. Analysis of the sequences of the mid-regions at locus 2 revealed an indel (Fig. 6C). The repetitive DNA flanking the indel is consistent with the pattern observed following strand slippage during DNA replication (10, 46).
FIG. 6.
Mid-regions of RD genes in four H. pylori isolates (corpus [two isolates], antrum, and fundus) from Ladakh subject 7. (A) Schematic of RD loci. The RD genotype of isolate 7C-sc3 differs from the other three isolates, with only a single RD gene at locus 1. (B) Schematic of mid-region organization. At locus 1, the mid-regions of three isolates were identical but differed from that of 7C-sc3. At locus 2, there were three different mid-region genotypes. (C) DNA sequence analysis. Repetitive DNA sequences on each side of the indel in locus 2 FAR2 gene mid-regions are indicated with purple lines.
Random amplified polymorphic DNA analysis was performed on the four H. pylori isolates from the patient represented in Fig. 6. Three of the isolates (7A-sc7, 7C-sc4, and 7F-sc1) showed identical patterns, indicating that they are clonally related, whereas the pattern for 7c-sc3 was completely different, indicating that this strain is different from the other three (data not shown). These results explain why the RD mid-region profile for 7C-sc3 was so different from those for the other three strains. Although the three related strains had identical mid-regions at locus 1, all three had different mid-regions at locus 2.
Multiple isolates from two additional patients also were examined (see Fig. S3 in the supplemental material). The mid-regions of the two isolates from the fundus of patient 30 differed by a single amino acid from the mid-regions of the five isolates from the corpus and fundus in the same patient (see Fig. S3A in the supplemental material). In addition to the nonsynonymous-nucleotide difference between the fundus isolates and the other isolates, the mid-regions of the isolates from the fundus also differed from the other isolates by two synonymous nucleotide changes. Eight isolates from patient 32 (two from the antrum, three from the corpus, and two from the fundus) had mid-regions identical at both the nucleotide and the amino acid levels (see Fig. S3B in the supplemental material). Thus, in an individual host, the dominant RD genotypes may be wholly, partially, or not at all conserved.
Human seroreactivity to recombinant JHP_0110 protein.
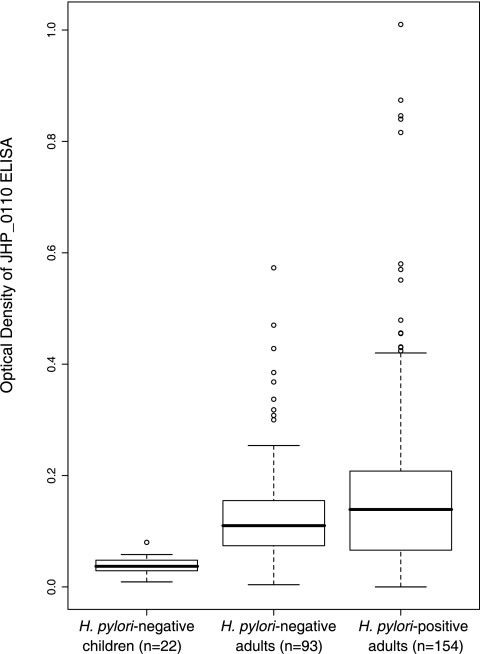
To determine whether the protein product of the RD gene JHP_0110 was recognized by H. pylori-colonized persons, ELISAs were conducted with recombinant JHP_0110 protein, using sera from H. pylori-positive adults and, as controls, from H. pylori-negative children and adults (Fig. 7). As expected, the sera from H. pylori-negative children (n = 22) showed little reactivity with the JHP_0110 protein (OD = 0.038 ± 0.017) (Fig. 7) and were used as a reference group. On the basis of this group, seropositivity was defined as >0.089 OD units (mean + 3 standard deviations). Both groups of adult subjects showed low-level reactivity, and the H. pylori-negative adults (OD = 0.138 ± 0.100; 66.6% seropositive) and the H. pylori-positive adults (OD = 0.179 ± 0.176; 63.0% seropositive) were not significantly different (P = 0.27).
FIG. 7.
Seroreactivity of H. pylori-negative children, H. pylori-negative adults, and H. pylori-positive adults to recombinant JHP_0110 protein. Box plots depict the median, interquartile range (box), and range. Outlier data points (values greater than 1.5 interquartile ranges from the interquartile range) are depicted by open circles. While sera from H. pylori-negative children showed very little reactivity (0.04 ± 0.02), sera from H. pylori-negative and H. pylori-positive adults were more reactive (0.14 ± 0.10 and 0.18 ± 0.18, respectively). The percents seroreactivity of the H. pylori-negative and H. pylori-positive adults were not significantly different (P = 0.27).
When Western blots of the recombinant JHP_0110 protein were probed with sera from 14 subjects, there was baseline reactivity for all, with no difference in band intensity between the H. pylori-positive sera and the H. pylori-negative sera. When JHP_0110 blots were exposed to the five sera most reactive under the ELISA conditions (Fig. 7), there also was little association between the IgG antibody level and the immunoblot findings (not shown). In total, these data do not support the hypothesis that the protein encoded by JHP_0110 is recognized as antigenic by H. pylori-positive subjects.
DISCUSSION
The previously recognized (19, 52) but not fully described group of genes that we now call the RD gene family was consistently observed at two well-defined genome loci in the 47 H. pylori strains that we studied. However, within these loci, we observed extensive variation at multiple levels: (i) the number of genes per strain; (ii) the identity of the FARs; (iii) the arrangement of genes at the two loci; and (iv) the composition, order, and length of the repetitive mid-regions. While we cannot be certain of the variation-generating mechanisms responsible for this diversity, the observed variation is likely related to the function of the proteins encoded by the RD gene family.
Variation in the number of RD genes across 47 strains and the phylogenetic relationships demonstrate that gene duplication in this family is common. While there were nearly equal numbers of RD genes at locus 1 and locus 2, the distribution by FAR type is nonrandom. This could reflect either functional considerations based on proximity of genes or founder effects. If the latter, it was most likely ancient, since the distribution is observed in isolates from widely dispersed parts of the world. Differing phylogenetic trees for the 5′ and 3′ regions of RD genes (Fig. 3) indicate that the opposite ends of each RD gene do not share a common evolutionary history. This finding could be explained by both inter- and intragenomic recombination, likely facilitated by the high level of identity shared by 3′ regions, FAR1s, FAR2s, and flanking genes.
The mid-regions of the RD genes exhibited the most intriguing variability, in the form of a complex “vocabulary” of heptad repeats. While the heptad amino acid “words” could be classified into 13 groups, the composition, order, and number of words varied greatly for the 19 RD genes in the five studied strains (Fig. 4). The variation in the number of repeats could provide encoded proteins with length-based functional differences, as has been observed for H. pylori FutA and FutB (50). However, it is also necessary to acknowledge the possibility that some of the repetitive DNA observed may be a result of repeated passing of cultures.
The specific mechanism behind the mid-region repeats is still unclear. The pattern observed in isolates obtained from individual hosts (Fig. 6; see also Fig. S3 in the supplemental material) is consistent with the hypothesis that repetitive DNA in the mid-region facilitates recombinatorial insertion or deletion events via slipped-strand mispairing (41, 46). However, imperfect repeats do not conform to expectations for slipped-strand repeat replication (13), nor is the evidence strong for gene conversion events in the mid-region. The presence of near-repeats suggests that many of the duplication events are very old and have not been subjected to gene conversion events occurring in concerted evolution, such as seen in rRNA genes (66) or in H. pylori babA and babB (58); their persistence suggests that each may represent a different functional allele. Thus, the observations described here may be an example of domain duplications (8, 75), which have been observed as genetic repeats from 11 to 609 nucleotides in length (42).
Domain duplications have been studied mostly for eukaryotic genomes (42, 75), and recombination (75) and selection at the protein level (35) are important in their evolution, but the mechanism for their origin is not understood. Repeats that become degenerate are more likely to be retained (23, 31, 75); our observation of words conserved between strains suggests selection for common function. The finding of the predominant words in the five studied strains among isolates from Ladakh, India, supports the notion of a common vocabulary in the H. pylori pan-genome. That the vocabulary has so many nuances suggests a repertoire capable of facilitating colonization in a wide variety of hosts in our outbred human population.
Tandem clusters of paralogous proteins often include membrane-associated proteins (33). Our finding of prokaryotic membrane lipoprotein lipid attachment sites in FAR1 sequences (Fig. 5) is consistent with the placement of these genes' protein products at the cell surface. A homologue of RD gene HP_1188 encodes a membrane-expressed protein that promotes attachment to gastric epithelial cells (64). When H. pylori cells were cocultured with AGS cells, HP_0118 expression was upregulated 2.9-fold and the encoded protein expressed at the cell surface (34). While an RD knockout showed in vitro growth characteristics indistinguishable from those of the wild type (19), its colonization efficiency was significantly impaired in a mouse model (52). In total, these experiments suggest that the RD genes encode adhesins that promote H. pylori attachment to host epithelial cells. The repetitive DNA in the RD gene family could be advantageous for colonization and persistence because domain duplication can lead to phenotypic variants and short-sequence DNA repeats facilitate adaptive characteristics, such as phase variation and antigenic variation (73).
How the extensive variability in RD genes is adaptive for H. pylori is not known. Phase variation would be unlikely since all observed repeats encoded 7 or 11 amino acids, but antigenic variation is still a possibility. Our observation that H. pylori-positive subjects are no more than minimally seroreactive to recombinant JHP_01110 suggests that this protein is not an H. pylori-specific antigen, that this protein is not reactive with human IgG under the conditions tested, that JHP_0110 is not expressed in vivo, or that mid-region hypervariability prevents the human immune system from mounting a full response. The last scenario would be similar to the observations for CagY, which also is a surface-exposed protein with extensive variation based on repetitive DNA (9, 18, 44). To fully substantiate this hypothesis, extensive testing with multiple RD antigens and various reaction conditions would have to be conducted.
That the mid-region is largely predicted to encode coiled-coil structures suggests functional possibilities; the ability of coiled-coil structures to dimerize (16, 47) increases the number of structural possibilities for the encoded RD proteins. With the most common genotype of single FAR1 and FAR2 genes, there are three possible dimers (FAR1/FAR1, FAR1/FAR2, and FAR2/FAR2). Since the RD genes within any strain are not identical and some strains have as many as five, the combinatorial dimer possibilities increase exponentially, providing a large reservoir of potential variants within each strain. In addition, coiled-coil structures may serve as pH-dependent molecular switches (16); pH-dependent conformational changes in adhesion could help adaptation to the acid gradient in the gastric lumen.
Expression of RD genes appears regulated by the two-component ArsRS system (HP_0166-HP_0165) (19), required for acid resistance in H. pylori (45). At pH ≤5, HP_0119 (48) and HP_1187 and HP_1188 (77) were all strongly upregulated, and expression levels of HP_0118, HP_0120, HP_1198, HP_1188, JHP_0110, and JHP_1113 were four- to eightfold greater in the wild type than in a ΔarsS strain (55). Alternative (but not exclusive) hypotheses are that low pH upregulates RD gene expression, promoting adhesion, enabling enhanced survival in the more nearly neutral paracellular niches, or that adhesion permits H. pylori to better regulate host gastric acid production (20).
Although our study did not identify the role of the RD gene family in H. pylori, our observations of conserved and diverse structures should provide a framework for future research. Since the RD proteins provide a competitive advantage to H. pylori during colonization (52) and we now document their genetic hypervariability, it is reasonable to conclude that the variability itself may be adaptive. Future studies of RD protein antigenicity will allow determination of whether their repetitive DNA facilitates antigenic or other functional variation. Molecular studies of protein-protein interactions in the RD gene family may uncover the functional significance of the two different FARs and the mid-region's coiled-coil structures.
Supplementary Material
Acknowledgments
This research was supported in part by R01 GM63270 and by the Diane Belfer Program in Human Microbial Ecology.
We thank David A. Baltrus and Karen Guillemin for sharing unpublished data, Ernst J. Kuipers for strains and information, and Edgardo L. Sanabria-Valentin for helpful discussions.
Footnotes
Published ahead of print on 11 September 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Achaz, G., E. P. Rocha, P. Netter, and E. Coissac. 2002. Origin and fate of repeats in bacteria. Nucleic Acids Res. 30:2987-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Achtman, M., T. Azuma, D. E. Berg, Y. Ito, G. Morelli, Z. J. Pan, S. Suerbaum, S. A. Thompson, A. van der Ende, and L. J. van Doorn. 1999. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol. Microbiol. 32:459-470. [DOI] [PubMed] [Google Scholar]
- 3.Aertsen, A., and C. W. Michiels. 2005. Diversify or die: generation of diversity in response to stress. Crit. Rev. Microbiol. 31:69-78. [DOI] [PubMed] [Google Scholar]
- 4.Akopyanz, N., N. O. Bukanov, T. U. Westblom, and D. E. Berg. 1992. PCR-based RFLP analysis of DNA sequence diversity in the gastric pathogen Helicobacter pylori. Nucleic Acids Res. 20:6221-6225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akopyanz, N., N. O. Bukanov, T. U. Westblom, S. Kresovich, and D. E. Berg. 1992. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 20:5137-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 7.Alm, R. A., and T. J. Trust. 1999. Analysis of the genetic diversity of Helicobacter pylori: the tale of two genomes. J. Mol. Med. 77:834-846. [DOI] [PubMed] [Google Scholar]
- 8.Apic, G., J. Gough, and S. A. Teichmann. 2001. Domain combinations in archaeal, eubacterial and eukaryotic proteomes. J. Mol. Biol. 310:311-325. [DOI] [PubMed] [Google Scholar]
- 9.Aras, R. A., W. Fischer, G. I. Perez-Perez, M. Crosatti, T. Ando, R. Haas, and M. J. Blaser. 2003. Plasticity of repetitive DNA sequences within a bacterial (type IV) secretion system component. J. Exp. Med. 198:1349-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aras, R. A., J. Kang, A. I. Tschumi, Y. Harasaki, and M. J. Blaser. 2003. Extensive repetitive DNA facilitates prokaryotic genome plasticity. Proc. Natl. Acad. Sci. USA 100:13579-13584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baltrus, D. A., M. R. Amieva, A. Covacci, T. M. Lowe, D. S. Merrell, K. M. Ottemann, M. Stein, N. R. Salama, and K. Guillemin. 2009. The complete genome sequence of Helicobacter pylori strain G27. J. Bacteriol. 191:447-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, and D. L. Wheeler. 2006. GenBank. Nucleic Acids Res. 34:D16-D20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bichara, M., J. Wagner, and I. B. Lambert. 2006. Mechanisms of tandem repeat instability in bacteria. Mutat. Res. 598:144-163. [DOI] [PubMed] [Google Scholar]
- 14.Bjorkholm, B., M. Sjolund, P. G. Falk, O. G. Berg, L. Engstrand, and D. I. Andersson. 2001. Mutation frequency and biological cost of antibiotic resistance in Helicobacter pylori. Proc. Natl. Acad. Sci. USA 98:14607-14612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blanchard, T. G., S. J. Czinn, and J. G. Nedrud. 1999. Host response and vaccine development to Helicobacter pylori infection. Curr. Top. Microbiol. Immunol. 241:181-213. [DOI] [PubMed] [Google Scholar]
- 16.Burkhard, P., J. Stetefeld, and S. V. Strelkov. 2001. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 11:82-88. [DOI] [PubMed] [Google Scholar]
- 17.Chedin, F., E. Dervyn, R. Dervyn, S. D. Ehrlich, and P. Noirot. 1994. Frequency of deletion formation decreases exponentially with distance between short direct repeats. Mol. Microbiol. 12:561-569. [DOI] [PubMed] [Google Scholar]
- 18.Delahay, R. M., G. D. Balkwill, K. A. Bunting, W. Edwards, J. C. Atherton, and M. S. Searle. 2008. The highly repetitive region of the Helicobacter pylori CagY protein comprises tandem arrays of an alpha-helical repeat module. J. Mol. Biol. 377:956-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dietz, P., G. Gerlach, and D. Beier. 2002. Identification of target genes regulated by the two-component system HP166-HP165 of Helicobacter pylori. J. Bacteriol. 184:350-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El-Omar, E. M., K. Oien, A. El-Nujumi, D. Gillen, A. Wirz, S. Dahill, C. Williams, J. E. Ardill, and K. E. McColl. 1997. Helicobacter pylori infection and chronic gastric acid hyposecretion. Gastroenterology 113:15-24. [DOI] [PubMed] [Google Scholar]
- 21.Felsenstein, J. 1985. Confidence-limits on phylogenies—an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 22.Finn, R. D., J. Mistry, B. Schuster-Bockler, S. Griffiths-Jones, V. Hollich, T. Lassmann, S. Moxon, M. Marshall, A. Khanna, R. Durbin, S. R. Eddy, E. L. Sonnhammer, and A. Bateman. 2006. Pfam: clans, web tools and services. Nucleic Acids Res. 34:D247-D251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Force, A., M. Lynch, F. B. Pickett, A. Amores, Y. L. Yan, and J. Postlethwait. 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151:1531-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Francois, F., J. Roper, A. J. Goodman, Z. Pei, M. Ghumman, M. Mourad, A. Z. de Perez, G. I. Perez-Perez, C. H. Tseng, and M. J. Blaser. 2008. The association of gastric leptin with oesophageal inflammation and metaplasia. Gut 57:16-24. [DOI] [PubMed] [Google Scholar]
- 25.Go, M. F., V. Kapur, D. Y. Graham, and J. M. Musser. 1996. Population genetic analysis of Helicobacter pylori by multilocus enzyme electrophoresis: extensive allelic diversity and recombinational population structure. J. Bacteriol. 178:3934-3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guruge, J. L., P. G. Falk, R. G. Lorenz, M. Dans, H. P. Wirth, M. J. Blaser, D. E. Berg, and J. I. Gordon. 1998. Epithelial attachment alters the outcome of Helicobacter pylori infection. Proc. Natl. Acad. Sci. USA 95:3925-3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Israel, D. A., N. Salama, U. Krishna, U. M. Rieger, J. C. Atherton, S. Falkow, and R. M. Peek, Jr. 2001. Helicobacter pylori genetic diversity within the gastric niche of a single human host. Proc. Natl. Acad. Sci. USA 98:14625-14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang, J., and M. J. Blaser. 2006. Bacterial populations as perfect gases: genomic integrity and diversification tensions in Helicobacter pylori. Nat. Rev. Microbiol. 4:826-836. [DOI] [PubMed] [Google Scholar]
- 29.Kang, J., and M. J. Blaser. 2008. Repair and antirepair DNA helicases in Helicobacter pylori. J. Bacteriol. 190:4218-4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang, J., D. Tavakoli, A. Tschumi, R. A. Aras, and M. J. Blaser. 2004. Effect of host species on recG phenotypes in Helicobacter pylori and Escherichia coli. J. Bacteriol. 186:7704-7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karev, G. P., Y. I. Wolf, A. Y. Rzhetsky, F. S. Berezovskaya, and E. V. Koonin. 2002. Birth and death of protein domains: a simple model of evolution explains power law behavior. BMC Evol. Biol. 2:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kersulyte, D., H. Chalkauskas, and D. E. Berg. 1999. Emergence of recombinant strains of Helicobacter pylori during human infection. Mol. Microbiol. 31:31-43. [DOI] [PubMed] [Google Scholar]
- 33.Kihara, D., and M. Kanehisa. 2000. Tandem clusters of membrane proteins in complete genome sequences. Genome Res. 10:731-743. [DOI] [PubMed] [Google Scholar]
- 34.Kim, N., E. A. Marcus, Y. Wen, D. L. Weeks, D. R. Scott, H. C. Jung, I. S. Song, and G. Sachs. 2004. Genes of Helicobacter pylori regulated by attachment to AGS cells. Infect. Immun. 72:2358-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kondrashov, F. A., and A. S. Kondrashov. 2006. Role of selection in fixation of gene duplications. J. Theor. Biol. 239:141-151. [DOI] [PubMed] [Google Scholar]
- 36.Kuipers, E. J., D. A. Israel, J. G. Kusters, M. M. Gerrits, J. Weel, A. van Der Ende, R. W. van Der Hulst, H. P. Wirth, J. Hook-Nikanne, S. A. Thompson, and M. J. Blaser. 2000. Quasispecies development of Helicobacter pylori observed in paired isolates obtained years apart from the same host. J. Infect. Dis. 181:273-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kulick, S., C. Moccia, X. Didelot, D. Falush, C. Kraft, and S. Suerbaum. 2008. Mosaic DNA imports with interspersions of recipient sequence after natural transformation of Helicobacter pylori. PLoS ONE 3:e3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 39.Kurtz, S., J. V. Choudhuri, E. Ohlebusch, C. Schleiermacher, J. Stoye, and R. Giegerich. 2001. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 29:4633-4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 41.Levinson, G., and G. A. Gutman. 1987. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol. Biol. Evol. 4:203-221. [DOI] [PubMed] [Google Scholar]
- 42.Li, W.-H. 1997. Molecular evolution. Sinauer Associates, Sunderland, MA.
- 43.Lin, E. A., X. S. Zhang, S. M. Levine, S. R. Gill, D. Falush, and M. J. Blaser. 2009. Natural transformation of Helicobacter pylori involves the integration of short DNA fragments interrupted by gaps of variable size. PLoS Pathog. 5:e1000337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu, G., T. K. McDaniel, S. Falkow, and S. Karlin. 1999. Sequence anomalies in the Cag7 gene of the Helicobacter pylori pathogenicity island. Proc. Natl. Acad. Sci. USA 96:7011-7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loh, J. T., and T. L. Cover. 2006. Requirement of histidine kinases HP0165 and HP1364 for acid resistance in Helicobacter pylori. Infect. Immun. 74:3052-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lovett, S. T. 2004. Encoded errors: mutations and rearrangements mediated by misalignment at repetitive DNA sequences. Mol. Microbiol. 52:1243-1253. [DOI] [PubMed] [Google Scholar]
- 47.Lupas, A. 1996. Coiled coils: new structures and new functions. Trends Biochem. Sci. 21:375-382. [PubMed] [Google Scholar]
- 48.Merrell, D. S., M. L. Goodrich, G. Otto, L. S. Tompkins, and S. Falkow. 2003. pH-regulated gene expression of the gastric pathogen Helicobacter pylori. Infect. Immun. 71:3529-3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moxon, R., C. Bayliss, and D. Hood. 2006. Bacterial contingency loci: the role of simple sequence DNA repeats in bacterial adaptation. Annu. Rev. Genet. 40:307-333. [DOI] [PubMed] [Google Scholar]
- 50.Nilsson, C., A. Skoglund, A. P. Moran, H. Annuk, L. Engstrand, and S. Normark. 2006. An enzymatic ruler modulates Lewis antigen glycosylation of Helicobacter pylori LPS during persistent infection. Proc. Natl. Acad. Sci. USA 103:2863-2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oh, J. D., H. Kling-Backhed, M. Giannakis, J. Xu, R. S. Fulton, L. A. Fulton, H. S. Cordum, C. Wang, G. Elliott, J. Edwards, E. R. Mardis, L. G. Engstrand, and J. I. Gordon. 2006. The complete genome sequence of a chronic atrophic gastritis Helicobacter pylori strain: evolution during disease progression. Proc. Natl. Acad. Sci. USA 103:9999-10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Panthel, K., P. Dietz, R. Haas, and D. Beier. 2003. Two-component systems of Helicobacter pylori contribute to virulence in a mouse infection model. Infect. Immun. 71:5381-5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peek, R. M., Jr., and M. J. Blaser. 2002. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat. Rev. Cancer 2:28-37. [DOI] [PubMed] [Google Scholar]
- 54.Pérez-Pérez, G. I., R. B. Sack, R. Reid, M. Santosham, J. Croll, and M. J. Blaser. 2003. Transient and persistent Helicobacter pylori colonization in Native American children. J. Clin. Microbiol. 41:2401-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pflock, M., N. Finsterer, B. Joseph, H. Mollenkopf, T. F. Meyer, and D. Beier. 2006. Characterization of the ArsRS regulon of Helicobacter pylori, involved in acid adaptation. J. Bacteriol. 188:3449-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pinto, A. V., A. Mathieu, S. Marsin, X. Veaute, L. Ielpi, A. Labigne, and J. P. Radicella. 2005. Suppression of homologous and homeologous recombination by the bacterial MutS2 protein. Mol. Cell 17:113-120. [DOI] [PubMed] [Google Scholar]
- 57.Pride, D. T., and M. J. Blaser. 2002. Concerted evolution between duplicated genetic elements in Helicobacter pylori. J. Mol. Biol. 316:629-642. [DOI] [PubMed] [Google Scholar]
- 58.Pride, D. T., R. J. Meinersmann, and M. J. Blaser. 2001. Allelic variation within Helicobacter pylori babA and babB. Infect. Immun. 69:1160-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rocha, E. P., A. Danchin, and A. Viari. 1999. Analysis of long repeats in bacterial genomes reveals alternative evolutionary mechanisms in Bacillus subtilis and other competent prokaryotes. Mol. Biol. Evol. 16:1219-1230. [DOI] [PubMed] [Google Scholar]
- 60.Rocha, E. P., A. Danchin, and A. Viari. 1999. Functional and evolutionary roles of long repeats in prokaryotes. Res. Microbiol. 150:725-733. [DOI] [PubMed] [Google Scholar]
- 61.Rohde, M., J. Puls, R. Buhrdorf, W. Fischer, and R. Haas. 2003. A novel sheathed surface organelle of the Helicobacter pylori cag type IV secretion system. Mol. Microbiol. 49:219-234. [DOI] [PubMed] [Google Scholar]
- 62.Romero, D., J. Martinez-Salazar, E. Ortiz, C. Rodriguez, and E. Valencia-Morales. 1999. Repeated sequences in bacterial chromosomes and plasmids: a glimpse from sequenced genomes. Res. Microbiol. 150:735-743. [DOI] [PubMed] [Google Scholar]
- 63.Romero-Gallo, J., G. I. Perez-Perez, R. P. Novick, P. Kamath, T. Norbu, and M. J. Blaser. 2002. Responses of endoscopy patients in Ladakh, India, to Helicobacter pylori whole-cell and Cag A antigens. Clin. Diagn. Lab. Immunol. 9:1313-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rubinsztein-Dunlop, S., B. Guy, L. Lissolo, and H. Fischer. 2005. Identification of two new Helicobacter pylori surface proteins involved in attachment to epithelial cell lines. J. Med. Microbiol. 54:427-434. [DOI] [PubMed] [Google Scholar]
- 65.Salama, N. R., G. Gonzalez-Valencia, B. Deatherage, F. Aviles-Jimenez, J. C. Atherton, D. Y. Graham, and J. Torres. 2007. Genetic analysis of Helicobacter pylori strain populations colonizing the stomach at different times postinfection. J. Bacteriol. 189:3834-3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Santoyo, G., and D. Romero. 2005. Gene conversion and concerted evolution in bacterial genomes. FEMS Microbiol. Rev. 29:169-183. [DOI] [PubMed] [Google Scholar]
- 67.Saunders, N. J., J. F. Peden, D. W. Hood, and E. R. Moxon. 1998. Simple sequence repeats in the Helicobacter pylori genome. Mol. Microbiol. 27:1091-1098. [DOI] [PubMed] [Google Scholar]
- 68.Sawyer, S. 1989. Statistical tests for detecting gene conversion. Mol. Biol. Evol. 6:526-538. [DOI] [PubMed] [Google Scholar]
- 69.Sawyer, S. A. 1999. GENECONV: a computer package for the statistical detection of gene conversion. Washington University, St. Louis, MO.
- 70.Suerbaum, S., and P. Michetti. 2002. Helicobacter pylori infection. N. Engl. J. Med. 347:1175-1186. [DOI] [PubMed] [Google Scholar]
- 71.Swofford, D. L. 2003. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4b10. Sinauer Associates, Sunderland, MA.
- 72.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 73.van Belkum, A., S. Scherer, L. van Alphen, and H. Verbrugh. 1998. Short-sequence DNA repeats in prokaryotic genomes. Microbiol. Mol. Biol. Rev. 62:275-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van der Ende, A., E. A. Rauws, M. Feller, C. J. Mulder, G. N. Tytgat, and J. Dankert. 1996. Heterogeneous Helicobacter pylori isolates from members of a family with a history of peptic ulcer disease. Gastroenterology 111:638-647. [DOI] [PubMed] [Google Scholar]
- 75.Vogel, C., S. A. Teichmann, and J. Pereira-Leal. 2005. The relationship between domain duplication and recombination. J. Mol. Biol. 346:355-365. [DOI] [PubMed] [Google Scholar]
- 76.Wang, G., M. Z. Humayun, and D. E. Taylor. 1999. Mutation as an origin of genetic variability in Helicobacter pylori. Trends Microbiol. 7:488-493. [DOI] [PubMed] [Google Scholar]
- 77.Wen, Y., E. A. Marcus, U. Matrubutham, M. A. Gleeson, D. R. Scott, and G. Sachs. 2003. Acid-adaptive genes of Helicobacter pylori. Infect. Immun. 71:5921-5939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wirth, H. P., M. Yang, R. M. Peek, Jr., J. Hook-Nikanne, M. Fried, and M. J. Blaser. 1999. Phenotypic diversity in Lewis expression of Helicobacter pylori isolates from the same host. J. Lab Clin. Med. 133:488-500. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.