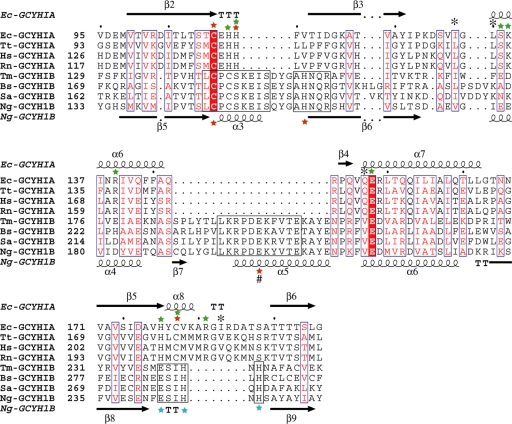
FIG. 7.
Structure-guided multisequence alignment of GCYH-IA and GCYH-IB in the shared T-fold region. For clarity, only four sequences from each subfamily are shown. Residue labels and secondary structure elements (nomenclature same as in Fig. 6) are shown above and below the sequence alignment for subfamilies A and B, respectively. Residues conserved between the two subfamilies are in blue boxes, with the invariant residues (the substrate-binding Glu and metal-coordinating Cys) highlighted in red. Conserved regions within the GCYH-IB subfamily are in black boxes. The metal ion liganding side chains in each subfamily are labeled with red stars. The # symbol indicates a metal liganding side chain from the neighboring subunit. Additional GCYH-IB-specific, strictly conserved active-site residues are labeled with blue stars. Residues known to interact via their side chains or backbones with the GTP substrate in crystal structures of GCYH-IA are labeled with green stars and black asterisks, respectively. Ec, Escherichia coli; Tt, Thermus thermophilus; Hs, Homo sapien; Rn, Rattus norvegicus; Tm, Thermotoga maritima; Bs, Bacillus subtilis; Sa, Staphylococcus aureus; Ng, Neisseria gonorrhoeae.