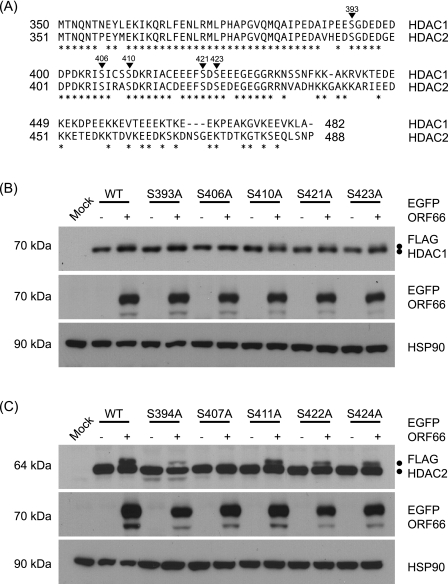
FIG. 5.
Identification of Ser residues in HDAC1 and HDAC2 that are phosphorylated in response to ORF66p expression. (A) Amino acid sequence alignment of the C termini of HDAC1 and HDAC2. The amino acid sequences of HDAC1 and HDAC2 are aligned, and positions marked with asterisks identify identical amino acids. Inverted triangles indicate phosphorylation sites in both proteins. The numbers above each triangle correspond to residues in HDAC1. (B) To map the ORF66p target phosphorylation site(s) in HDAC1, 293A cells were mock transformed or cotransformed with 1 μg of either pFLAG-HDAC1, pFLAG-S393A, pFLAG-S406A, pFLAG-S410A, pFLAG-S421A, or pFLAG-S423A and 1 μg of pEGFP-C1 or pEGFP-ORF66. After 48 h, cells were harvested and equal amounts of total protein (20 μg) from each sample were analyzed by Western blotting using antisera specific for FLAG, GFP, and HSP90. (C) To map the ORF66p target phosphorylation site(s) in HDAC2, 293A cells were mock transformed or cotransformed with 1 μg of pFLAG-HDAC2, pFLAG-S394A, pFLAG-S407A, pFLAG-S411A, pFLAG-S422A, or pFLAG-S424A and 1 μg of pEGFP or pEGFP-ORF66. After 48 h, cells were harvested and equal amounts of total protein (20 μg) from each sample were analyzed by Western blotting using antisera specific for FLAG, GFP, and HSP90.