Abstract
To elucidate the epigenetic regulation of Tat-independent human immunodeficiency virus (HIV) transcription following proviral integration, we constructed an HIV type 1 (HIV-1)-based replication-defective viral vector that expresses a reporter green fluorescent protein (GFP) product from its intact long terminal repeat (LTR). We transduced this construct into human tumor cell lines that were either deficient in or competent for the Brm-type SWI/SNF complex. One day after transduction, single cells that expressed GFP were sorted, and the GFP expression profiles originating from each of these clones were analyzed. Unlike clones of the SWI/SNF-competent cell line, which exhibited clear unimodal expression patterns in all cases, many clones originating from Brm-deficient cell lines either showed a broad-range distribution of GFP expression or were fully silenced. The resorting of GFP-negative populations of these isolated clones showed that GFP silencing is either reversible or irreversible depending upon the proviral integration sites. We further observed that even in these silenced clones, proviral gene transcription initiates to accumulate short transcripts of around 60 bases in length, but no elongation occurs. We found that this termination is caused by tightly closed nucleosome-1 (nuc-1) at the 5′ LTR. Also, nuc-1 is remodeled by exogenous Brm in some integrants. From these results, we propose that Brm is required for the occasional transcriptional elongation of the HIV-1 provirus in the absence of Tat. Since the Brm-type SWI/SNF complex is expressed at marginal levels in resting CD4+ T cells and is drastically induced upon CD4+ T-cell activation, we speculate that it plays crucial roles in the early Tat-independent phase of HIV transcription in affected patients.
Human immunodeficiency virus type 1 (HIV-1) proviral DNA is semirandomly integrated into the host cell genome. The transcription of the HIV-1 provirus is characterized by an early Tat-independent phase and a late Tat-dependent phase. In the early Tat-independent phase, HIV-1 transcription is dependent upon the interaction of host transcription factors with cis-regulatory DNA elements with the viral 5′ long terminal repeat (LTR) (23, 26) and the assembly of the transcription apparatus including RNA polymerase II (RNAPII) on these sequences. Importantly, RNAPII synthesizes mostly abortive transcripts during this Tat-independent phase (13, 16). Only a small fraction of the HIV-1 transcripts are in fact expected to be elongated and produce the transactivator Tat protein. Tat and its cellular coactivator, positive transcription factor b (pTEFb), bind the transacting responsive (TAR) element present in the 5′ region of the HIV-1 proviral transcript and cause the hyperphosphorylation of RNAPII, which then elongates this transcript (20, 31). Hence, for the successful elucidation of HIV-1 expression, host factors involved in the first stage of HIV-1 expression will be very important, although the low expression levels of these HIV-1 transcripts may make a detailed analysis difficult.
The SWI/SNF chromatin remodeling complex plays many important roles in epigenetic regulation in human cells. This complex contains either Brm or BRG1 as the catalytic subunit but not both (30). Each of these proteins possesses DNA-dependent ATPase activity, and although they have significant overlapping functions, they do show distinct roles in some biological activities. For example, we previously reported that the Brm-type but not the BRG1-type SWI/SNF complex is required for the stable gene transcription of murine leukemia virus (MLV)-based retrovirus vectors (18) as well as cdx2-dependent villin expression in gastrointestinal cells (36). In Brm-deficient cells, we found that single cells that were transduced with a replication-defective MLV-based vector carrying the lacZ reporter frequently formed colonies containing a mixture of LacZ-positive (LacZ+) and LacZ-negative (LacZ−) cells (mosaic colonies). A chromatin immunoprecipitation (ChIP) assay showed that histone deacetylase 1 (HDAC1), HDAC2, and YY1 are recruited around the MLV 5′ LTR when the cells are deficient in Brm (18). It is currently not known whether this requirement of Brm for stable MLV expression is also true for other retroviruses.
In our current study, we examined whether the Brm-type SWI/SNF complex is required for stable HIV-1 transcription in the absence of the Tat protein. Using HIV-1-based replication-defective viral vectors, we show that HIV-1 proviral gene expression is destabilized in Brm-deficient cell lines and further that this expression is very often silenced promptly and, in some integrants, can even be reversibly reactivated. We also show that this resulted not from blocks at the level of transcriptional initiation but rather from the suppression of transcriptional elongation. We provide evidence that the Brm-type SWI/SNF chromatin remodeling complex is involved in the disruption of nucleosome-1 (nuc-1), which is present just downstream of the transcription start site, in order to promote the efficient elongation of HIV-1 transcripts.
MATERIALS AND METHODS
Cell culture and preparation of PBMCs.
The human cell lines SW13(vim−) (38), C33A, HeLa-S3, HeLa-S3-shBrm4 (37), H1299, AZ521, A204, and 293FT (Invitrogen) were incubated in Dulbecco's modified Eagle's medium with 10% fetal calf serum (Life Technologies/Invitrogen) at 37°C. Peripheral blood mononuclear cells (PBMCs) were obtained from a single healthy donor, purified on Ficoll gradients, and then stained with fluorescein isothiocyanate-conjugated anti-CD4 and phycoerythrin-conjugated anti-HLA-DR antibodies (eBioscience). Monocytes were purified from PBMC cultures by sorting for lymphocytes with dull CD4 and then seeded for 2 days. Proteins were extracted from adherent cells. Resting T cells were purified from PBMCs by sorting for lymphocytes with a high level of expression of CD4 but low levels of HLA-DR. Activated CD4+ T cells were collected as follows: specific antibodies were immobilized on 48-well tissue culture plates by incubating 20 μg/ml anti-CD3 (UCHT1) (eBioscience) and 10 μg/ml anti-CD28 (CD28.2) (eBioscience) in phosphate-buffered saline overnight at 4°C. Excess antibodies were removed from the wells by washing with phosphate-buffered saline. Resting CD4+ T cells were added at 2.5 × 106 cells/well in 0.5 ml of complete RPMI 1640 medium containing 10% fetal calf serum and incubated at 37°C and 5% CO2. Proteins were collected at 2 days and 6 days after activation. This study was approved by the institutional ethical review board for human investigation at the Institute of Medical Science, University of Tokyo, Tokyo, Japan (approval number 19-27-0131).
Cell sorting and GFP analysis.
Cell sorting and flow cytometric analyses were performed using FACSAria and FACSCalibur apparatuses (Becton Dickinson), respectively. For green fluorescent protein (GFP) fluorescence analysis, dead cells were eliminated on the basis of their forward and side scattering using propidium iodide fluorescence.
Plasmids and DNA transfection.
The 0.7-kb XhoI-StuI PCR fragment of the NL432 HIV-1 molecular clone (encoding the polypurine tract and the 3′ LTR) (1) was inserted into pLenti6/V5-D-TOPO (Invitrogen) to generate pWL-X. The 3.3-kb BamHI fragment of nlsLacZ (4) and the 0.7-kb fragment of GFP were inserted into the ClaI-XhoI site of pWL-X to generate the constructs pWL-LacZ and pWL-GFP (pWLG), respectively. Plasmids pCAGF1-IRES-Kusabira-Orange (pCAGF1-IKO) (empty vector), pCAGF1-IKO-Brm, and pCAGF1-IKO-Brm (KR) were generated by inserting the 0.7-kb Kusabira Orange fragment of phKO1-MC1 (MBL) into the SmaI-EcoRV site of pCAGF1-IG, pCAGF1-IG-Brm, and pCAGF1-IG-Brm (KR), respectively (37). Oligonucleotides pairs (5′-TTTGGGTCTCTCTGGTTAGACCAGATCTGAGCCTGGGAGCTCTCTGGCTAACTAGGGAACCCACTTTTTTTG-3′ and 5′-AATTCAAAAAAAGTGGGTTCCCTAGTTAGCCAGAGAGCTCCCAGGCTCAGATCTGGTCTAACCAGAGAGACC-3′) were annealed and ligated into vector pmU6 (11) to generate pmU6-62nt. These expression plasmids were transfected into human cell lines using FugeneHD (Roche).
Vector production.
Vesicular stomatitis virus G protein-pseudotyped lentiviral vectors (pWL-LacZ and pWL-GFP) were produced using the Virapower lentiviral expression system (Invitrogen).
Mosaic colony assay.
A mosaic colony assay was performed essentially as described previously (18). Briefly, cells were transduced with pWL-LacZ at a low multiplicity of infection (MOI) (less than 0.2). At 1 day after transduction, the transduced cultures were trypsinized and seeded at a very low cell density to enable single-colony formation on the plates. Three days after seeding, the cells were assessed for LacZ expression. pCAGF1-IKO and pCAGF1-IKO-Brm were transduced into C33A cells, and 24 h after transfection, Kusabira Orange-positive cells were sorted 1 day before performing the mosaic colony assay.
Genomic and inverse PCRs.
Genomic DNAs were extracted from pWLG-transduced cell cultures, and 0.1 μg of these DNA extracts was then used for genomic PCR using the same primer pairs and amplification conditions as those for the ChIP protocol described below. For inverse PCR, genomic DNAs were digested with EcoRI or PstI at 37°C overnight and subjected to self-ligation at 16°C overnight. This facilitated the generation of self-ligated circular DNA carrying both a 5′-terminal proviral fragment (from the 5′ LTR to GFP) and its neighboring genomic sequences. PCR and nested PCR using the following primer sets were then performed to amplify DNA fragments encompassing vector and cellular DNA junctions: primers GFP101 (5′-GTCCGCCCTGAGCAAAGA-3′) and A2 (5′-CAAAGGTCAGTGGATATCTG-3′) for the first PCR and primers A4 (5′-CGATCACATGGTCCTGCTGG-3′) and A3 (5′-CAATCAGGGAAGTAGCCTTG-3′) for nested PCR. Amplified fragments were subjected to sequencing analysis and human genome mapping using BLAST searches.
RT-PCR.
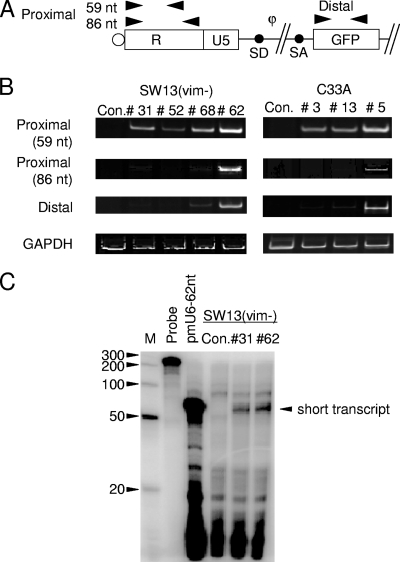
Total RNA was prepared from cells using the mirVana microRNA isolation kit (Ambion). Total RNA was treated with DNase (Takara) in accordance with the manufacturer's instructions. Semiquantitative reverse transcription-PCR (RT-PCR) was performed using a Superscript one-step reaction with the Platinum Taq kit (Invitrogen). The primer sets used were as follows: start-F (5′-GGGTCTCTCTGGTTAGA-3′) and short-R1 (5′-GGGTTCCCTAGTTAGCC-3′) for the proximal 59 nucleotides (nt), start-F and short-R2 (5′-GGCAAGCTTTATTGAG-3′) for the proximal 86 nt, 5′-TACCGGTCGCCACCATGGTGA-3′ and 5′-GTACTCCAGCTTGTGCCCCAG-3′ for GFP, and 5′-ACCACAGTCCATGCCATCAC-3′ and 5′-TCCACCACCCTGTTGCTGTA-3′ for GAPDH (glyceraldehyde-3-phosphate dehydrogenase). The band intensities were semiquantified by densitometry (Atto Printgraph).
Western blotting.
Western blotting was performed as described previously (18) by using antibodies against Brm (Abcam), BRG1 (Santa Cruz Biotechnology), BAF155 (Santa Cruz Biotechnology), BAF60a (BD Transduction Laboratories), HIV-1 p24 (39/5.4A; Abcam), and β-actin (BD Transduction Laboratories).
RNase protection assay.
A small RNA fraction (less than 200 nt) was prepared from cells using the mirVana microRNA isolation kit (Ambion). The 32P-labeled HIV-1-specific RNA probe (positions −117 to +86 of the LTR and a noncomplementary stretch of 32 nt at the 5′ end) was prepared by in vitro transcription of the antisense probe using T7 RNA polymerase (Invitrogen). Fifteen micrograms of these cellular small RNAs was hybridized overnight with 32P-labeled RNA probes (6 × 105 cpm) at 37°C. An RNase protection assay was performed using the RPAIII kit (Ambion). Protected fragments were electrophoresed on 20% polyacrylamide gels at a high temperature. 32P-end-labeled DynaMarker RNA Low II (BioDynamics Laboratory) RNA markers were also loaded as size standards.
ChIP assay.
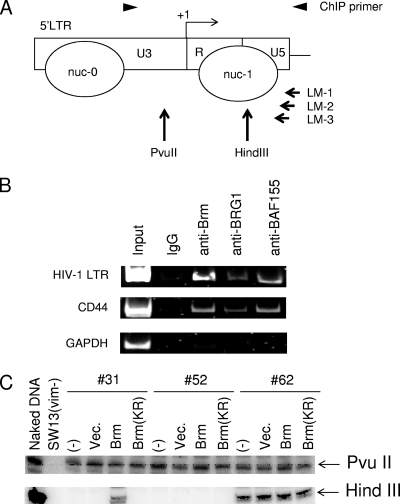
ChIP assays were performed in accordance with the manufacturer's protocol for the ChIP assay kit (Upstate Biotechnology). The specific antibodies used for immunoprecipitation were anti-Brm (Abcam), anti-BRG1 (Santa Cruz Biotechnology), anti-BAF155 (Santa Cruz Biotechnology), and anti-normal rabbit immunoglobulin G (Santa Cruz Biotechnology). The amplification conditions used for semiquantitative PCR were as follows: 30 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s. The primers used were as follows: LTR-F (5′-GAGCTTTCTACAAGGGACTTTCCG-3′) and LTR-R (5′-CCGTGCGCGCTTCAG-3′) (384 bp), CD44-F (5′-TTCGGTCATCCTCTGTCCTGACG-3′) and CD44-R (5′-AATGAGGCTGCCTCGGAAGTTG-3′) (345 bp), and GAPDH-F (5′-TGACTGTCGAACAGGAGGAG-3′) and GAPDH-R (5′-GCTACTAGCGGTTTTACGGG-3′) (174 bp). PCR products were visualized using SYBR green I staining after 5 and 10% polyacrylamide gel electrophoresis. The band intensities were semiquantified by densitometry using the LAS4000UV minisystem (Fuji Film).
Restriction enzyme accessibility assays and ligation-mediated PCR.
SW13(vim−) clones were harvested and resuspended at 2.5 × 106 cells/ml in cold buffer A (10 mM Tris-HCl [pH 7.4], 10 mM NaCl, 3 mM MgCl2, and 0.3 M sucrose) for 5 min and then lysed in 0.3% NP-40 for 3 min. Cellular nuclei were collected by centrifugation (1,500 rpm at 4°C), resuspended in buffer B (10 mM Tris-HCl [pH 7.9], 10 mM MgCl2, 50 mM NaCl, 1 mM dithiothreitol, 100 μg/ml bovine serum albumin, and 0.1 mM phenylmethylsulfonyl fluoride), and digested with 4 U of HindIII and PvuII for 16 h. Genomic DNA was extracted from nuclei with TES buffer (10 mM Tris [pH 8.0], 400 mM NaCl, 2 mM EDTA, and 1% sodium dodecyl sulfate) and treatment with 200 μg/ml proteinase K at 55°C for 5 h. DNA was extracted once with a 1× volume of phenol, followed by a 1:1 phenol-chloroform-isoamyl alcohol extraction and two extractions with 24:1 chloroform-isoamyl alcohol. The ligation-mediated PCR primer used was exactly the same as that described previously (39).
Quantitation of HIV-1 viral production by TNF-α stimulation.
ACH-2 and ACH-2-derived cells (1 × 106 cells/24-well plate) were stimulated by TNF-α (10 ng/ml) (R&D Systems) in the presence of 50 μM azidothymidine; culture supernatants were then collected by centrifugation at 0, 8, 12, or 24 h poststimulation; and viral production was monitored by HIV-1 p24 antigen enzyme-linked immunosorbent assay (ELISA) (ZeptoMetrix) according to the manufacturer's instructions. The cellular pellets were used for analysis of the cellular p24 protein by Western blotting.
RESULTS
HIV-1 vectors exhibit destabilized reporter expression in Brm-deficient human tumor cell lines.
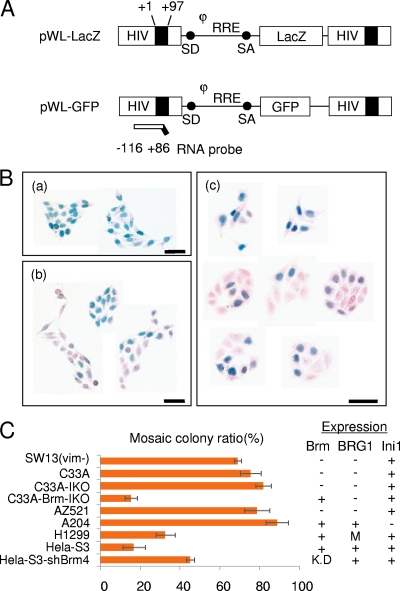
We previously reported, using a mosaic colony assay, that the Brm-type SWI/SNF complex is required for the maintenance of the stable gene expression of MLV-based retrovirus vectors (18). To determine whether this complex is also necessary for the stable transcription of HIV-1 in the absence of Tat, we analyzed the stability of HIV-1 proviral transcription using the same assay. First, we constructed an HIV-1-based LacZ reporter vector under the control of the intact HIV-1 LTR (pWL-LacZ) (Fig. 1A) and transduced this into either SWI/SNF component-competent or Brm-deficient human cells at a low MOI (less than 0.2) to minimize the introduction of multiple proviral copies into a single cell. The transduced cell cultures were grown for 1 day to obtain complete cellular segregation and were then seeded at a low density to enable single-colony formation. The colonies that formed 3 days after seeding were stained for LacZ to assess reporter expression levels (Fig. 1B). When the HIV-1 LacZ vector was transduced into an SWI/SNF-competent cell line such as HeLa-S3, the cells formed colonies in which all the progeny cells expressed LacZ (positive colonies), similar to MLV-based vectors (Fig. 1Ba) (18). However, most Brm-deficient colonies were composed of mixed populations of both LacZ+ and LacZ− cells (mosaic colonies) (Fig. 1Bc). When the mosaic colony ratios (the mosaic colony ratio was calculated by dividing the mosaic colony number by the sum of the mosaic colony number and the positive colony number) for several human tumor cell lines were determined, Brm-deficient cell lines such as SW13(vim−), C33A, and AZ521 exhibited a high ratio. In cell lines competent for the Brm-type SWI/SNF complex, such as HeLa-S3 and H1299 (deficient in BRG1), the mosaic colony ratio was low, as expected, whereas HeLa-S3 cells in which Brm is knocked down (HeLa-S3-shBrm4) showed a higher mosaic colony ratio than the parental HeLa-S3 cells (Fig. 1Bb and C). On the other hand, C33A cells transiently introduced with Brm clearly exhibited a much lower mosaic colony ratio than did those transfected with a control vector (Fig. 1C). A204 cells, which are deficient in Ini1, a core component of the SWI/SNF complex (21), also exhibited a high mosaic colony ratio. These results indicate that the Brm- but not the BRG1-type SWI/SNF complex is required for HIV-1 proviral gene expression in the absence of Tat.
FIG. 1.
Prompt silencing of HIV-1-based vector expression occurs in some human cell lines. (A) Schematic representation of vectors pWL-LacZ and pWL-GFP. The position of the RNA probe (nt −116 to +86) used for RNase protection analysis is shown. The vectors contain an intact LTR, from which the lacZ or GFP reporter gene is driven from the HIV LTR. SD and SA, splice donor and splice acceptor sites, respectively; RRE, Rev-responsive element, which is necessary to export full-length proviral mRNA from the nucleus to the cytoplasm; φ, packaging signal that is necessary for the packaging of full-length vector RNA into the viral particle. (B) Cells were transduced with vector pWL-LacZ at a low MOI (less than 0.2) to minimize the introduction of multiple proviral copies. At 1 day after transduction, the transduced cultures were trypsinized and seeded at a very low cell density to enable single-colony formation. In vector-transduced HeLa-S3 cells (a), positive colonies were mostly observed. HeLa-S3-shBrm4 (b) and C33A (c) cells transduced with the pWL-LacZ vector formed mosaic colonies, as evidenced by the LacZ expression pattern on day 3 after seeding. (C) The mosaic colony ratio was calculated by dividing the mosaic colony number by the sum of the mosaic colony number and the positive colony number. The expression statuses for Brm, BRG1, and Ini1 are summarized at right. K.D and M represent “knockdown” and “mutation,” respectively. The error bars indicate the standard deviations (n = 3).
Clonal analysis of HIV-1 LTR expression in Brm-deficient cell cultures.
Although a mosaic colony assay using pWL-LacZ is both a convenient and sensitive method for detecting gene silencing within a few days, it cannot sufficiently analyze clonal differences between the integrants in detail or the reversibility of the silencing. To overcome these limitations, the lacZ gene was replaced with the GFP gene in the expression viral vector to yield pWLG (GFP reporter vector) (Fig. 1A), which was then introduced into the Brm-deficient cell lines SW13(vim−) and C33A and the SWI/SNF-competent cell line HeLa-S3. At 1 day after this transduction, single cells expressing GFP were sorted by flow cytometry and then grown for 3 weeks to obtain clonal cultures (see Fig. S1 in the supplemental material).
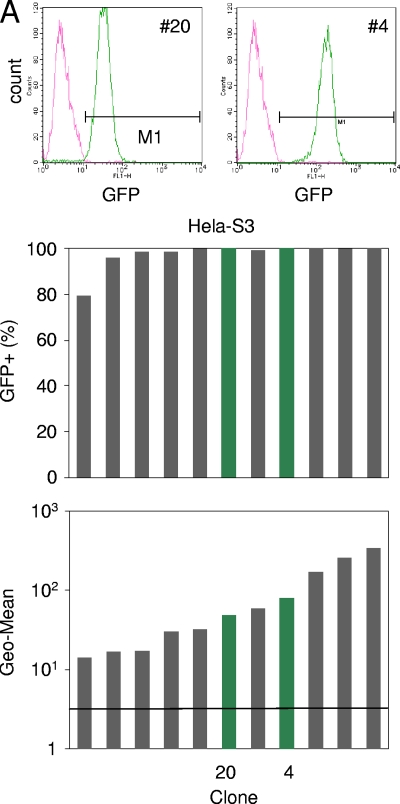
We obtained 137 clonal cultures: 43 SW13(vim−), 83 C33A, and 11 HeLa-S3 transductants. All of these clones were confirmed to harbor at least one proviral copy by genomic PCR, indicating that cells expressing GFP at least 1 day after transduction were successfully cloned. Among these 137 clones, 46 were randomly selected and further characterized for the copy number of the integrated provirus. Southern blot analysis demonstrated that most clones (average, 84.8%) [16/17 SW13(vim−), 15/21 C33A, and 8/8 HeLa-S3 clones] contained no more than one proviral DNA copy. Since we transduced the GFP reporter vector into cells at an MOI of 0.2 in this experiment, about 9% of the clonal cultures would be predicted to contain more than two integrants using a Poisson distribution of transduction for the entire cellular population (5). This indicated that the experimental results are consistent with theoretical predictions. Although a small degree of contamination by multicopy integrants is unavoidable in this type of experiment, we contend that this does not significantly affect the GFP expression profiles. The representative clones HeLa-S3 clone 4 (HeLa-S3-4); HeLa-S3-20; SW13(vim−)-31, -52, -62, and -78; and C33A-3, -5, -13, and -25, which were used in our closed analysis, however, were confirmed to harbor a single provirus.
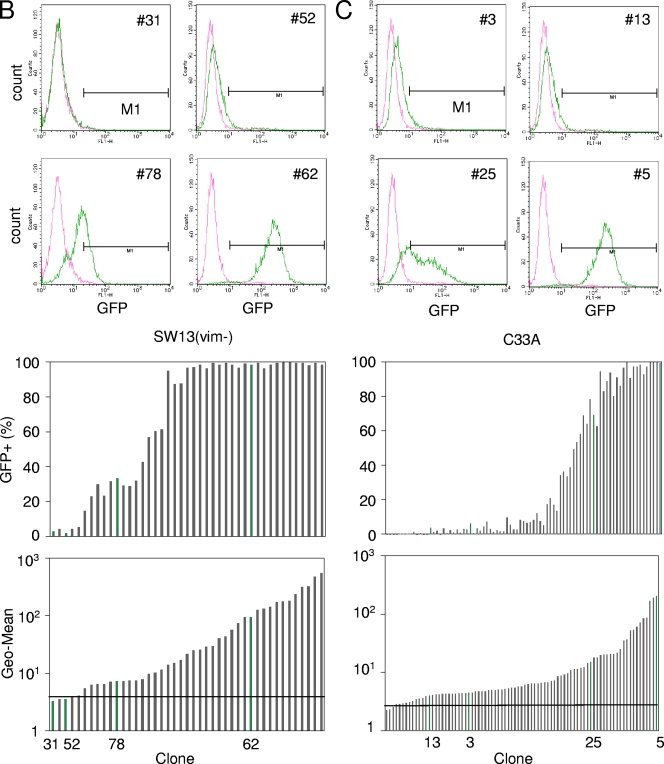
All of the transductant clones of the SWI/SNF-competent HeLa-S3 cell line exhibited clear unimodal expression patterns (examples include HeLa-S3-20 and -4) (Fig. 2A, top). Among clones of Brm-deficient cell lines (Fig. 2B and C, top), we found that there were several clones that were fully silenced [SW13(vim−)-31 and -52 and C33A-3 and -13) and many clones for which the GFP expression level ranged broadly from GFP negative (GFP−) to clearly GFP positive (GFP+) [variegated clones, SW13(vim−)-78 and C33A-25]. These tendencies were shown clearly when we plotted the percentages of GFP+ cells in each clone (Fig. 2, middle) and geometrical means of the GFP expression levels of the entire clonal population (Fig. 2, bottom). Here, we expediently defined GFP+ cells as those that have a GFP level higher than that of autofluorescence (M1 gate). These variegated expression profiles observed for Brm-deficient cellular clones are consistent with our observations of mosaic colonies in the LacZ vector-transduced clones (Fig. 1B).
FIG. 2.
Single-cell sorting of HIV vector-transduced cells. Vector pWL-GFP was transduced into human cancer cell lines at a low MOI (0.2). At 24 h after transduction, GFP+ cells were single-cell sorted into 96-well plates (FACSAria cell sorting system). Clonal cell lines were analyzed for their GFP expression profiles at 3 weeks after sorting, and fluorescence-activated cell sorter patterns of some representative clones are shown (upper fluorescence-activated cell sorter analysis data and green bar in bar graph). The pink line and green line represent parental cell lines and sorted clones, respectively. The middle bar graph indicates the percentage of GFP+ cells, which were defined as cells within the M1 bar. The bottom bar graph indicates the geometric means (Geo-Mean) of cellular clones of HeLa-S3 (A), SW13(vim−) (B), and C33A (C). The crossbars represent the geometric means of the parental cells (autofluorescence). In these bar graphs, the sorted clones were ordered using geometric means.
We further analyzed the GFP expression profiles of H1299 cells, which are BRG1 deficient but have intact Brm, and A204 cells, which lack Ini1, an integral component of the SWI/SNF complex. Like HeLa-S3 cells, all of the 42 clones of H1299 cells isolated here showed clear unimodal expression patterns (see Fig. S2 in the supplemental material), whereas A204 clones frequently showed variegated expression of GFP in a manner similar to that of SW13(vim−) and C33A (data not shown). These results indicate that Tat-independent HIV-1 expression is destabilized in the absence of the Brm-type SWI/SNF complex.
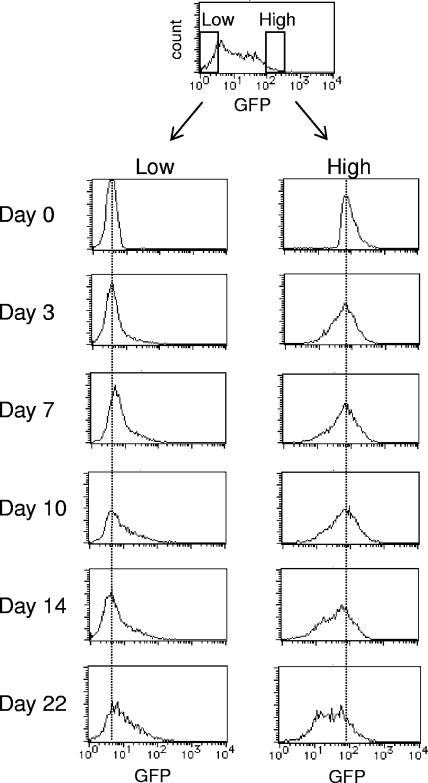
To examine the fluctuation of GFP expression in clones such as C33A-25, which shows typical variegated expression, we sorted the GFP− and GFP+ subpopulations in cultures of C33A-25 cells. By following the time course of the GFP expression patterns in each of these sorted subpopulations, we found that some GFP− cells became GFP+ and vice versa and that after 3 weeks, each culture showed a broad GFP expression profile that was similar to those of the parental clones (Fig. 3). These results suggest that silencing is reversible in the clones with a broad GFP expression profile. On the other hand, none of the fully silenced clones of SW13(vim−) (clones 31 and 52) or C33A (clones 3 and 13) reexpressed GFP after subsequent growth, indicating that this silencing is irreversible.
FIG. 3.
Time course analysis of the resorting of an HIV vector-transduced clone, C33A-25. Cell fractions from the C33A-25 cellular populations with either low or high GFP expression levels were resorted and grown for about 3 weeks. GFP expression profiles of the aliquots were analyzed on the days indicated.
We next examined whether GFP expression levels of C33A-25 cells have some correlation with the cell cycle or not. Clone 25 cells were sorted, and their GFP levels and DNA contents (DyeCycle Violet) were simultaneously determined. We compared the GFP expression profile of cells in the G0/G1 phase to that of cells in the G2/M phase. Their GFP expression profiles were very similar, which were also similar to that of the original clone 25 culture (see Fig. S3 in the supplemental material). This result shows that GFP expression levels are not affected by the cell cycle. It was previously reported that CpG methylation could be a mechanism to maintain HIV-1 latency in long-term-infected U937 cells (25). We therefore examined the status of DNA methylation in the LTR region of these cellular clones by using a bisulfate sequencing method. We observed only very a rare and marginal CpG methylation profile in these silenced and variegated clones except for SW13(vim−)-52 cells (see Fig. S4 in the supplemental material). It is notable that both the GFP− and GFP+ subpopulations in cultures of C33A-25 cells were not methylated like their parental C33A-25 cells. These results indicate that in most cases, our observed silenced or variegated expressions of GFP were not caused by CpG methylation around the HIV promoter region.
By using wild-type HIV-1 and an HIV-1-based vector, it was previously shown that HIV-1 preferentially integrates within introns of coding genes in both human T-cell lines and cancer cell lines and that 60 to 70% of the proviruses integrate into transcription units (24, 35). To elucidate the integration sites in our current sorted clones, we examined the integration sites of several clones of SW13(vim−), C33A, and A204 transductants by inverse PCR and found that most of their integration sites were within the introns of coding genes (Table 1), as was reported previously (24, 35). It is also noteworthy that even though clones such as SW13(vim−)-52 and -76 and C33A-3 and -13 were irreversibly silenced, the proviruses did not reside in heterochromatin. Also, at the same time, we determined the transcriptional orientation of the integrated proviral DNA compared with the host gene where this proviral DNA had integrated but found no obvious causal association (Table 1).
TABLE 1.
Chromosomal features associated with the integration sites of HIV-1 vector pWLG in cellular clones
| Clone | GFP expression profile | Chromosomal locus | Location of integration site (gene name) | Provirus direction vs host gene |
|---|---|---|---|---|
| SW13(vim−)-31 | Silenced | —a | ||
| SW13(vim−)-43 | Broad | Chr6q22.31 | Intronic (RNF217) | Reverse |
| SW13(vim−)-49 | Expressing | Chr5q21 | Intergenic | |
| SW13(vim−)-52 | Silenced | Chr.2p13 | Intronic (ZNF638) | Reverse |
| SW13(vim−)-63 | Broad | Chr9p24.1 | Intronic (NFIB) | Forward |
| SW13(vim−)-68 | Expressing | Chr.9q32 | Intronic (KIAA1958) | Reverse |
| SW13(vim−)-76 | Silenced | Chr.1q41 | Intronic (C1orf80) | Forward |
| SW13(vim−)-77 | Expressing | Chr.21q22.3 | Intronic (PCNT) | Reverse |
| SW13(vim−)-78 | Broad | Chr.16p13.3 | Intergenic | |
| C33A-3 | Silenced | Chr.11p11.2 | Intronic (C11orf49) | Reverse |
| C33A-13 | Silenced | Chr.11q13 | Intronic (DPH4) | Forward |
| A204-7 | Broad | Chr.1p36.1 | Intergenic |
—, Integrated into a highly repetitive sequence. The exact locus cannot be identified.
HIV-1 transcripts are not elongated under Brm-deficient conditions.
To gain further insight into the role of Brm in HIV-1 transcription, we wished to examine the transcriptional steps during HIV-1 promoter transcription that were restricted in Brm-deficient cells. It was reported previously that only a small fraction of the initial HIV-1 transcripts are fully elongated in the absence of Tat. Whereas the major transcripts produced in this early phase seem to be prematurely terminated, this would eventually lead to an accumulation of Tat and a trigger of drastic late-phase transcription (13, 17). Hence, we first examined whether transcription is initiated in GFP-silenced Brm-deficient clones. Transcriptional initiation and elongation of the HIV-1 vector were assayed by RT-PCR using primer pairs that cover the transcriptional start site and the GFP gene (Fig. 4A). Although no amplified band corresponding to the GFP gene (the distal portion of transcript) was detected in GFP-silenced SW13(vim−)-31 and -52 and C33A-3 and -13 cells, an amplified proximal-region band of 59 nt was clearly evident in these silenced clones, including SW13(vim−)-52, whose proviral DNA was methylated (Fig. 4B). By a comparison of the PCR bands, more amplification was detected using a primer set of 59 nt than 86 nt in the GFP-silenced clones SW13(vim−)-31 and -52 and C33A-3 and -13 (Fig. 4B). These results suggest that short transcripts had ceased elongation at between 60 and 86 nt after initiation in these GFP-silenced clones. This indicates that the major barrier to gene silencing in Brm-deficient cells does not occur at the transcriptional initiation step because a premature termination of proviral gene transcription is clearly detectable. To detect intact short transcripts of 60 to 86 nt, we employed an RNase protection assay and analyzed some sorted clones. We designed an RNA probe complementary to the region between positions −117 and +86 of the LTR (the transcription start site is at position +1) and containing a noncomplementary 32-nt sequence at its 5′ end (Fig. 1A). As a size control for these short transcripts, we constructed an expression cassette for the 62-nt TAR element stem-loop region of HIV-1 transcripts driven by a mouse polymerase III-type U6 promoter (pmU6-62nt) and transduced this cassette into 293FT cells. For the detection of short transcripts, we collected small RNA fractions (under 200 nt) and performed an RNase protection assay with these clones. As shown in Fig. 4C, we observed a distinct single protected band of about 60 nt in the GFP+ clone SW13(vim−)-6 and in the GFP− clone SW13(vim−)-31. This result shows that this short transcript had been generated shortly after the TAR stem-loop structure in GFP− clones.
FIG. 4.
Analysis of RNA transcripts in the pWLG-transduced cell clones. (A) Schematic of the pWL-GFP vector showing the location of RT-PCR primers (arrowheads). The proximal primers amplify both abortive and full-length products (R region of the 5′ LTR and the 3′ LTR) of transcription. The distal primer amplifies any full-length product of transcription. SD and SA, splice donor and splice acceptor sites, respectively. (B) Expression levels of the proviral transcripts were measured in the clones SW13(vim−)-31, -52, -68, and -62 and C33A-3, -13, and -5 as well as the parental cell lines SW13(vim−) and C33A by semiquantitative RT-PCR. Con., control. (C) RNase protection analysis of the clones SW13(vim−)-31 and -62. The RNA probe used is described in the legend of Fig. 1A. Fifteen micrograms of small RNA extracts was used and hybridized with this probe at 37°C. Protected RNA fragments were resolved using 20% denaturing acrylamide gel electrophoresis at a high temperature. As a positive control, 62 nt of the TAR stem-loop region of HIV-1 transcripts driven by a mouse polymerase III type U6 promoter (pmU6-62nt) was used.
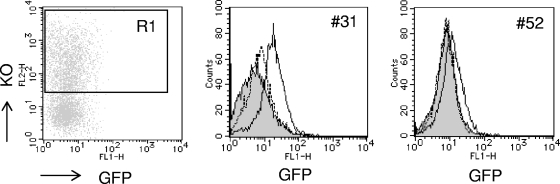
To further examine the role of Brm in Tat-independent HIV-1 proviral transcription, we transfected a Brm, Brm-KR (a dominant negative Brm), or empty vector into the GFP-silenced clones (see Fig. S5A in the supplemental material). As shown in Fig. 5 and Fig. S5B in the supplemental material, we observed a low-level but clear induction of GFP expression in nearly the entire population of SW13(vim−)-31 cells and some other clones only when transfected with wild-type Brm, indicating that the ATPase activity of Brm is essential for leaky transcriptional elongation to occur in the absence of Tat. In contrast, the induction of GFP expression was not observed in clones such as SW13(vim−)-52 when transfected with wild-type Brm. Interestingly, clone 52 was shown to be a rare clone, where frequent CpG methylation was detected around the proviral 5′ LTR region (see Fig. S4 in the supplemental material). These results suggest that the responsiveness to Brm is dependent upon each specific proviral integration site and further that several kinds of epigenetic chromatin modifications could have accumulated in the promoter during each cellular expansion after GFP+ cell sorting.
FIG. 5.
Exogenous Brm transduction of GFP-silenced clones. SW13(vim−) clones were transfected with vectors carrying Brm and Brm-KR as well as a control plasmid (con). At 2 days after transfection, KO-positive cells (R1 gated) were sorted, and their GFP expression profiles were analyzed by flow cytometry. Brm-transduced cells are indicated by the bold line, Brm-KR-transduced cells are indicated by the dotted line, and nontransduced parental clones are indicated by the gray line.
Brm-type SWI/SNF is recruited to the HIV-1 promoter region and alters the chromatin structure around the HIV-1 promoter.
We next conducted a ChIP assay to examine whether the Brm-type SWI/SNF complex is recruited to the HIV-1 promoter in vivo. The 5′ LTR portion of HIV-1 proviral DNA was amplified by PCR (Fig. 6A). As the positive control for this assay system, we used the CD44 promoter region where SWI/SNF complexes are recruited, and as a negative control, we utilized the GAPDH promoter region at which AP-1 and SWI/SNF complexes are not recruited (Fig. 6B) (9). In this assay, we used the GFP-expressing clone HeLa-S3-4, which harbors a single proviral copy. We found from this analysis that Brm was recruited around the HIV-1 promoter site. Consistently, it has been shown that BAF155, which has been reported to be a core component for the SWI/SNF complex, is also recruited around the HIV-1 promoter site (21). These results suggest that Brm will be recruited to the HIV-1 promoter as an SWI/SNF complex. We found that the alternative catalytic subunit of the SWI/SNF complex, BRG1, is also recruited to the HIV-1 promoter, as was seen in CD44 promoter region. Whereas we cannot currently explain why recruited BRG1 does not seem to activate the transcription of this promoter, a similar selective activation potential by the Brm-type SWI/SNF complex was observed previously for the villin promoter (36).
FIG. 6.
Analysis of the HIV-1 LTR chromatin structure. (A) Map of the restriction sites and positions of nucleosomes (nuc-0 and nuc-1) in the HIV-1 LTR. The positions of ligation-mediated (LM) PCR primers are shown. The arrowheads indicate the PCR primer set used for the ChIP assay. (B) ChIP assay around the HIV-1 promoter site in the HeLa-S3-4 clone. Normal rabbit immunoglobulin G (IgG) was used as an antibody control. The PCR products were separated by polyacrylamide gel electrophoresis and stained with SYBR green I. Analysis of the GAPDH promoter was performed as a negative control, and analysis of the CD44 promoter was performed as a positive control. (C) Restriction enzyme accessibility assay of the HIV-1 LTR. Nuclei were isolated from untreated clones SW13(vim−)-31, -52 and -62 and transfected with vectors expressing Brm, Brm-KR, or a control vector (Vec.). As a positive control, naked genomic DNA was purified from SW13(vim−)-52 and then digested with PvuII or HindIII.
The chromatin structure and the accessibility of a promoter region to host transcriptional factors are important for successful transcription. Importantly, it was previously reported that the displacement of a positioned nuc-1 downstream of a transcriptional start site is a key process for the successful transcriptional elongation of the HIV-1 provirus (28, 29). Hence, to assess the status of nuc-1 in our current analyses, we conducted a restriction enzyme accessibility assay using some of our selected GFP+ and GFP− SW13(vim−) clones. After the isolated nuclei were separately treated with the restriction enzymes shown in Fig. 6A, DNA was purified from them, and a primer extension reaction was performed using LM-1 as the primer (Fig. 6A). After linker ligation, PCR was performed using the sequence complementary to the linker and LM-2 as the primer pair. The degree of digestion was finally measured by a primer extension reaction using 32P-labeled LM-3 as the primer (ligation-mediated PCR). In all of the GFP+ and GFP− clones analyzed herein, the PvuII site present outside of the positioned nucleosomes was equally susceptible to digestion by this enzyme. However, the HindIII site that is present in nuc-1 was much more resistant to HindIII digestion in GFP− clones [SW13(vim−)-31 and -52] than the GFP+ clone SW13(vim−)-62 (Fig. 6C). These results indicate that the nuc-1 structure of the GFP− clones in Brm-deficient cells is more rigid. Next, we tested whether these rigid nuc-1 structures are released following the induction of exogenous Brm. We transfected the Brm expression plasmid into SW13(vim−) clones harboring provirus, and 48 h after transfection, we harvested the nuclei from transfected cells and assessed the nuc-1 structure using a restriction enzyme accessibility assay. The extent of digestion of the HindIII site in SW13(vim−)-62 and -52 cells was unaltered by the exogenous transduction of Brm and Brm-KR. In contrast, the digestion of the HindIII site in SW13(vim−)-31 was increased only when exogenous Brm was induced. These data are consistent with data from our flow cytometric analysis showing that GFP expression levels were unchanged when exogenous Brm was transduced into the clones SW13(vim−)-62 and -52 (Fig. 5 and see Fig. S5B in the supplemental material). These results provide evidence that the Brm-type SWI/SNF complex is often successfully disrupting the nuc-1 structure to support transcriptional elongation and also that the extent of the reliance of HIV-1 provirus elongation upon Brm-type SWI/SNF is dependent upon the integration site of this provirus.
Brm expression levels are strongly suppressed in resting CD4+ T cells.
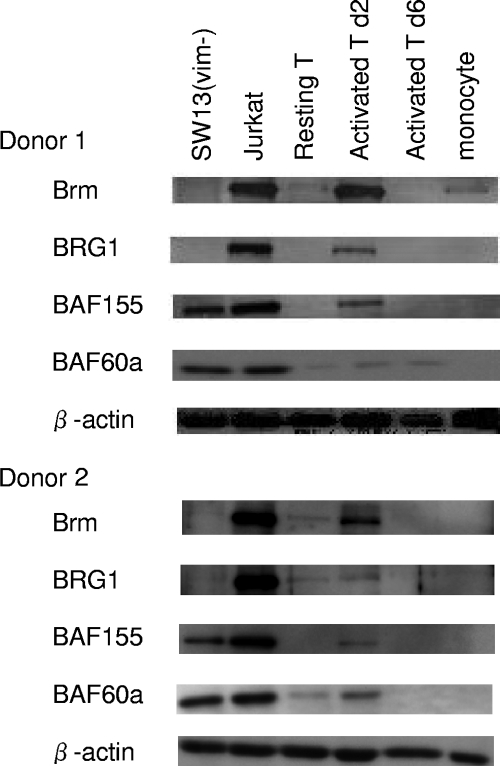
To determine the status of the SWI/SNF complex in natural host cells of HIV-1, we analyzed the expression levels of Brm and of other components of this complex in human T cells and monocytes. Resting CD4+ T cells from PBMCs from healthy donors were purified by sorting the populations that expressed high levels of CD4 but no HLA-DR, a representative activation marker, whereas monocytes were purified from a CD4-dull fraction (see Fig. S6 in the supplemental material). One-half of the resting CD4+ T cells obtained were further cultured in the presence of anti-CD3 and anti-CD28 to obtain activated T cells. By Western blotting (Fig. 7), it was found that the expression levels of the Brm, BRG1, BAF60a, and BAF155 subunits of the SWI/SNF complex in resting CD4+ T cells were very low in two healthy donors. Importantly, activated CD4+ T cells transiently induced Brm and other subunits of the SWI/SNF complex at 2 days after activation, and these subunits were then downregulated at 6 days after activation. In the case of monocytes, all of the subunits of the SWI/SNF complex examined were at marginal levels. For HIV patients undergoing highly active antiretroviral therapy (HAART) treatments, it was previously reported that cells latently infected with HIV-1 are detected principally in resting CD4+ T cells that had been infected during the transition from an activated to a resting state (10). Our current result suggests the possibility that Brm plays important roles in the early Tat-independent phase of HIV-1 transcription in reactivated CD4+ T cells in these patients.
FIG. 7.
Purification and characterization of resting CD4+ T cells from a healthy donor. Shown is Western blotting of components of the SWI/SNF complex in resting CD4+ T cells, activated CD4+ T cells, and some human cancer cell lines. Brm and other components of SWI/SNF complexes were immunoblotted at 0, 2, and 6 days after cellular activation with anti-CD3 and anti-CD28 monoclonal antibodies.
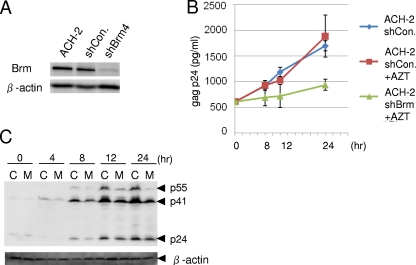
Brm knockdown suppresses HIV production in ACH-2 cells.
To examine whether the Brm protein is required for the reactivation of HIV-1 proviral expression from lymphocytic cells latently infected with HIV-1, we used the ACH-2 cell line as latently infected model cells. The ACH-2 cell line was derived from a T-lymphocytic parental cell line (CEM) that was latently infected with intact HIV and has been used extensively as model cell system for HIV latency and activation (22). In unstimulated ACH-2 cells, only a marginal basal level of RNA synthesis is detected, which consists predominantly of multiply spliced transcripts coding for regulatory proteins (22). Since ACH-2 cells express the Brm protein, we transduced cells with a short hairpin RNA against Brm (shBrm) expression retroviral vector into ACH-2 cells and established stable transductants by drug selection. The expression level of Brm was significantly reduced in ACH-2 cells transduced with the shBrm vector compared with those in cells transduced with a control vector (Fig. 8A). After stimulation with TNF-α, we examined HIV production in the culture medium from these transduced ACH-2 cells. To minimize reinfection after TNF-α stimulation, we added azidothymidine, an RT inhibitor, into medium prior to TNF-α stimulation. When p24 ELISA was performed, levels of HIV particles were not different between parental ACH-2 cells and cells expressing control short hairpin RNA (shRNA). On the other hand, the production of HIV-1 particles from shBrm-transduced ACH-2 cells showed significantly suppressed production compared with that of ACH-2 cells expressing control shRNA during 24 h after TNF-α stimulation (Fig. 8B).
FIG. 8.
Brm is required for the induction of HIV production from ACH-2 cells stimulated with TNF-α. (A) Expression levels of Brm in parental ACH-2 cells or those transduced with a retroviral vector expressing shBrm or control shRNA (shCon.). β-Actin was used for the internal control. (B) Kinetics of HIV production from these ACH-2 cells. Culture media were collected by centrifugation at 0, 8, 12, or 24 h after TNF-α (10 ng/ml) stimulation. p24gag antigen was detected by ELISA for the quantification of HIV particles. AZT, azidothymidine. (C) Expression of gag proteins in ACH-2 cells expressing shBrm (M) or control shRNA (C). Cellular pellets obtained in B were analyzed by Western blotting using anti-p24 antiserum.
We also determined the expression levels of endogenous gag proteins by Western blotting using anti-p24 antiserum. Although the accumulated amounts of the p24gag protein are only slightly lower in cells expressing shBrm, the expression levels of its precursor, p55, were clearly lower in shBrm-transduced ACH-2 cells than in those expressing control shRNA (Fig. 8C). These results also support that Brm is necessary to reactivate proviral expression from latently infected ACH-2 cells.
DISCUSSION
To examine whether the Brm-type SWI/SNF complex is required for reporter gene transcription from the HIV-1 LTR in the absence of the Tat protein, we analyzed cellular clones transduced with HIV-1-based vectors carrying either LacZ or GFP (Fig. 1A). As was previously seen in MLV-based vector transduction experiments, we observed that LacZ reporter expression from an HIV-1-based vector was promptly and specifically silenced in Brm-deficient human tumor cell lines and that transduced cellular clones derived from these populations frequently formed mosaic colonies (mixture of both LacZ− and LacZ+ cells) (Fig. 1B). Although the mosaic colony assay using the LacZ reporter gene is a convenient and sensitive method for detecting gene silencing, it is not sufficient to analyze either the clonal differences between the integrants in detail or the reversibility of these silencing events. When single cells transduced with GFP vectors were sorted for GFP+ cells at 1 day after transduction and clonally expanded, we were able to assess the proviral gene expression profile of each integrant. Notably, in Brm-deficient cells, we frequently found GFP-silenced clones and also variegated clones that formed broad GFP expression patterns. Importantly, also, a subpopulation of GFP− cells in this clonal culture reexpressed GFP, suggesting that silencing is reversible in cellular clones that show such broad GFP expression profiles.
It is very significant that silencing mechanisms in Brm-deficient cell lines appear to differ between MLV-based and HIV-1-based vectors. We previously reported that the silencing of an MLV-based vector in Brm-deficient cells was at the transcriptional initiation level and is caused by the recruitment of YY-1 and HDAC1/2 to the LTR region. In our current study, we have found that silencing is due mainly to the strong blockage of elongation as a result of a Brm deficiency (Fig. 4). Our analysis by RT-PCR (Fig. 4B) and RNase protection assays (Fig. 4C) reveals that proviral transcription is abortively terminated at around 60 nt in GFP-silenced clones. Here, however, we cannot exclude the possibility that the short transcript was a product of being processed from longer transcripts by digestion with exonucleases from their 3′ ends. We also show that inefficient elongation is at least partly caused by a failure in the displacement of nuc-1 downstream of the transcriptional start site of the 5′ LTR. Unlike the situation for GFP+ clones, nuc-1 was not remodeled in GFP− clones such as SW13(vim−)-31 (Fig. 6C). We found that the introduction of exogenous Brm into these cells leads to a recovery of GFP expression and that this is associated with the efficient remodeling of nuc-1 in transduced clones. In several other GFP-silenced clones analyzed by flow cytometry, the GFP expression levels were also found to be recovered to various degrees by the transduction of exogenous Brm (see Fig. S5B in the supplemental material). These results suggest that Brm is required for the displacement of nuc-1 during proviral elongation in the absence of Tat. The different recovery levels of GFP expression in each integrant may be due to epigenetic changes around the promoter region that had accumulated during the course of cellular cloning over 3 weeks. By use of ChIP analysis, we demonstrated that Brm is clearly recruited to the HIV-1 LTR in a GFP-expressing HeLa-S3 transductant clone (Fig. 6B). Since BAF155 is also recruited to the HIV-1 LTR, Brm is probably recruited to the promoter as a part of an SWI/SNF complex. We also found that BRG1 is recruited to the HIV-1 LTR. Given that LacZ and GFP expression levels were stable in a BRG1-deficient cell line, H1299 (Fig. 1C and see Fig. S2 in the supplemental material), we think that the recruited BRG1 would not have a significant transactivating function at the HIV-1 LTR in the absence of Tat.
It was reported previously that latent HIV-1 proviruses are present in resting CD4+ T cells of patients undergoing HAART (14). It is further believed that HIV-1 is unable to replicate efficiently in resting memory T cells because there are several viral infection and replication blocks in these cells (10). Hence, latent infection would need to have been established in resting memory T cells originating from activated T cells that were infected with HIV-1 during the process of their inactivation. Whereas latently infected cells produce little HIV-1 proviral mRNA in patients with HAART (2, 3, 7, 12), it was previously reported that abortive transcripts of about 60 nt are detectable in resting T cells from AIDS patients undergoing HAART (14). Interestingly, we show from our present data that Brm and some other components of the SWI/SNF complex, which are marginal in resting CD4+ T cells, are transiently induced when they are activated by CD3 and CD28 costimulation (Fig. 7). These observations suggest that the lack of expression of the Brm-type SWI/SNF complex would be one of the major causes of the attenuated HIV-1 proviral expression observed in resting CD4+ T cells.
In the presence of Tat, stochastic expression patterns of HIV were observed previously by several groups (8, 19, 32-34). For example, it was previously reported by Weinberger et al. that Tat stochastics appear to be sufficient to generate phenotypic bifurcation (32). Here, it is assumed that there is an initial or preexisting Tat concentration of 5 to 50 molecules in the cell, which is necessary to generate phenotypic bifurcation according to predictions in silico. In our study, on the other hand, stochastic gene expression from the HIV LTR was observed in the complete absence of Tat. Weinberger et al. analyzed the expression of an LTR-GFP control HIV vector (LG) that does not produce Tat at all and concluded that their GFP levels in Jurkat cells (competent for Brm) are low and stable, which is basically consistent with data from our studies using HeLa-S3 and H1299 cell lines, which are also competent for Brm. Importantly, when host cells lacked Brm, we always observed frequent variegation in HIV LTR expression in the absence of Tat. Because the natural HIV host cells in latent infection, resting CD4+ T cells, contain marginal levels of Brm (Fig. 7) and also because HIV production from a T-lymphocyte cell line latently infected with HIV, ACH-2, was strongly suppressed by knocking down Brm (Fig. 8), our observations reveal that a lack of Brm would limit and fluctuate proviral RNA elongation and further that HIV expression would be variegated in this earliest stage in these host cells. Such stochastic fluctuations in HIV expression in the absence of Tat would be further amplified by a Tat-mediated positive transcriptional feedback loop (32-34) after the Tat protein has been sufficiently accumulated. Concerning this transcriptional regulation in the earliest stage, there are important analyses of cis elements (8) and the status of histone modification (19) in the HIV LTR that were reported previously. In our current analysis, the Brm-type SWI/SNF complex was shown to be involved mainly in transcriptional elongation. However, it is possible that this complex would have some contribution to the regulation of transcriptional initiation together with host factors such as SP-1 (8) and HP-1-alpha as well as histone-modifying enzymes (19).
Interestingly, it was previously reported that the Tat-mediated activation of the HIV-1 promoter requires the SWI/SNF complex as a coactivator (6, 15, 27). Our current data indicated that the Brm-type SWI/SNF complex is necessary for long-transcript synthesis in the absence of Tat. These observations indicate that SWI/SNF plays essential roles in HIV-1 infection in both an early Tat-independent phase and a late Tat-dependent phase and further reveal that an understanding of the dynamism of the components and pathways involving the SWI/SNF complex during the HIV-1 infection cycle will be important to achieve in future projects.
Supplementary Material
Acknowledgments
We thank Akio Adachi for providing the NL432 HIV-1 plasmid and also Sayoko Kawaura and Aki Kato for assistance in preparing the manuscript. We thank the IMSUT FACS Core Laboratory for cell sorting.
This work was supported by a grant-in-aid for scientific research on priority areas and grants by the Program of Founding Research Center for Emerging and Reemerging Infectious Disease, funded by the contract research fund from the Ministry of Health, Labor, and Welfare and the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan. T.M. is a recipient of funding from PRESTO.
Footnotes
Published ahead of print on 2 September 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams, M., L. Sharmeen, J. Kimpton, J. M. Romeo, J. V. Garcia, B. M. Peterlin, M. Groudine, and M. Emerman. 1994. Cellular latency in human immunodeficiency virus-infected individuals with high CD4 levels can be detected by the presence of promoter-proximal transcripts. Proc. Natl. Acad. Sci. USA 91:3862-3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adams, M., C. Wong, D. Wang, and J. Romeo. 1999. Limitation of Tat-associated transcriptional processivity in HIV-infected PBMC. Virology 257:397-405. [DOI] [PubMed] [Google Scholar]
- 4.Arai, T., K. Matsumoto, K. Saitoh, M. Ui, T. Ito, M. Murakami, Y. Kanegae, I. Saito, F. L. Cosset, Y. Takeuchi, and H. Iba. 1998. A new system for stringent, high-titer vesicular stomatitis virus G protein-pseudotyped retrovirus vector induction by introduction of Cre recombinase into stable prepackaging cell lines. J. Virol. 72:1115-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arai, T., M. Takada, M. Ui, and H. Iba. 1999. Dose-dependent transduction of vesicular stomatitis virus G protein-pseudotyped retrovirus vector into human solid tumor cell lines and murine fibroblasts. Virology 260:109-115. [DOI] [PubMed] [Google Scholar]
- 6.Ariumi, Y., F. Serhan, P. Turelli, A. Telenti, and D. Trono. 2006. The integrase interactor 1 (INI1) proteins facilitate Tat-mediated human immunodeficiency virus type 1 transcription. Retrovirology 3:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brooks, R., M. J. Rotheram-Borus, E. G. Bing, G. Ayala, and C. L. Henry. 2003. HIV and AIDS among men of color who have sex with men and men of color who have sex with men and women: an epidemiological profile. AIDS Educ. Prev. 15:1-6. [DOI] [PubMed] [Google Scholar]
- 8.Burnett, J. C., K. Miller-Jensen, P. S. Shah, A. P. Arkin, and D. V. Schaffer. 2009. Control of stochastic gene expression by host factors at the HIV promoter. PLoS Pathog. 5:e1000260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujita, S., T. Ito, T. Mizutani, S. Minoguchi, N. Yamamichi, K. Sakurai, and H. Iba. 2008. miR-21 gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J. Mol. Biol. 378:492-504. [DOI] [PubMed] [Google Scholar]
- 10.Han, Y., K. Lassen, D. Monie, A. R. Sedaghat, S. Shimoji, X. Liu, T. C. Pierson, J. B. Margolick, R. F. Siliciano, and J. D. Siliciano. 2004. Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected individuals carry integrated HIV-1 genomes within actively transcribed host genes. J. Virol. 78:6122-6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haraguchi, T., T. Mizutani, N. Yamamichi, T. Ito, S. Minoguchi, and H. Iba. 2007. siRNAs do not induce RNA-dependent transcriptional silencing of retrovirus in human cells. FEBS Lett. 581:4949-4954. [DOI] [PubMed] [Google Scholar]
- 12.Hermankova, M., J. D. Siliciano, Y. Zhou, D. Monie, K. Chadwick, J. B. Margolick, T. C. Quinn, and R. F. Siliciano. 2003. Analysis of human immunodeficiency virus type 1 gene expression in latently infected resting CD4+ T lymphocytes in vivo. J. Virol. 77:7383-7392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kao, S. Y., A. F. Calman, P. A. Luciw, and B. M. Peterlin. 1987. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature 330:489-493. [DOI] [PubMed] [Google Scholar]
- 14.Lassen, K. G., J. R. Bailey, and R. F. Siliciano. 2004. Analysis of human immunodeficiency virus type 1 transcriptional elongation in resting CD4+ T cells in vivo. J. Virol. 78:9105-9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahmoudi, T., M. Parra, R. G. Vries, S. E. Kauder, C. P. Verrijzer, M. Ott, and E. Verdin. 2006. The SWI/SNF chromatin-remodeling complex is a cofactor for Tat transactivation of the HIV promoter. J. Biol. Chem. 281:19960-19968. [DOI] [PubMed] [Google Scholar]
- 16.Marciniak, R. A., B. J. Calnan, A. D. Frankel, and P. A. Sharp. 1990. HIV-1 Tat protein trans-activates transcription in vitro. Cell 63:791-802. [DOI] [PubMed] [Google Scholar]
- 17.Marciniak, R. A., and P. A. Sharp. 1991. HIV-1 Tat protein promotes formation of more-processive elongation complexes. EMBO J. 10:4189-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mizutani, T., T. Ito, M. Nishina, N. Yamamichi, A. Watanabe, and H. Iba. 2002. Maintenance of integrated proviral gene expression requires Brm, a catalytic subunit of SWI/SNF complex. J. Biol. Chem. 277:15859-15864. [DOI] [PubMed] [Google Scholar]
- 19.Pearson, R., Y. K. Kim, J. Hokello, K. Lassen, J. Friedman, M. Tyagi, and J. Karn. 2008. Epigenetic silencing of human immunodeficiency virus (HIV) transcription by formation of restrictive chromatin structures at the viral long terminal repeat drives the progressive entry of HIV into latency. J. Virol. 82:12291-12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peterlin, B. M., and D. H. Price. 2006. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 23:297-305. [DOI] [PubMed] [Google Scholar]
- 21.Phelan, M. L., S. Sif, G. J. Narlikar, and R. E. Kingston. 1999. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell 3:247-253. [DOI] [PubMed] [Google Scholar]
- 22.Pomerantz, R. J., D. Trono, M. B. Feinberg, and D. Baltimore. 1990. Cells nonproductively infected with HIV-1 exhibit an aberrant pattern of viral RNA expression: a molecular model for latency. Cell 61:1271-1276. [DOI] [PubMed] [Google Scholar]
- 23.Roebuck, K. A., and M. Saifuddin. 1999. Regulation of HIV-1 transcription. Gene Expr. 8:67-84. [PMC free article] [PubMed] [Google Scholar]
- 24.Schroder, A. R., P. Shinn, H. Chen, C. Berry, J. R. Ecker, and F. Bushman. 2002. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110:521-529. [DOI] [PubMed] [Google Scholar]
- 25.Singh, M. K., and C. D. Pauza. 1992. Extrachromosomal human immunodeficiency virus type 1 sequences are methylated in latently infected U937 cells. Virology 188:451-458. [DOI] [PubMed] [Google Scholar]
- 26.Stevens, M., E. De Clercq, and J. Balzarini. 2006. The regulation of HIV-1 transcription: molecular targets for chemotherapeutic intervention. Med. Res. Rev. 26:595-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Treand, C., I. du Chene, V. Bres, R. Kiernan, R. Benarous, M. Benkirane, and S. Emiliani. 2006. Requirement for SWI/SNF chromatin-remodeling complex in Tat-mediated activation of the HIV-1 promoter. EMBO J. 25:1690-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Lint, C., S. Emiliani, M. Ott, and E. Verdin. 1996. Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J. 15:1112-1120. [PMC free article] [PubMed] [Google Scholar]
- 29.Verdin, E., P. Paras, Jr., and C. Van Lint. 1993. Chromatin disruption in the promoter of human immunodeficiency virus type 1 during transcriptional activation. EMBO J. 12:3249-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, W., Y. Xue, S. Zhou, A. Kuo, B. R. Cairns, and G. R. Crabtree. 1996. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 10:2117-2130. [DOI] [PubMed] [Google Scholar]
- 31.Wei, P., M. E. Garber, S. M. Fang, W. H. Fischer, and K. A. Jones. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92:451-462. [DOI] [PubMed] [Google Scholar]
- 32.Weinberger, L. S., J. C. Burnett, J. E. Toettcher, A. P. Arkin, and D. V. Schaffer. 2005. Stochastic gene expression in a lentiviral positive-feedback loop: HIV-1 Tat fluctuations drive phenotypic diversity. Cell 122:169-182. [DOI] [PubMed] [Google Scholar]
- 33.Weinberger, L. S., R. D. Dar, and M. L. Simpson. 2008. Transient-mediated fate determination in a transcriptional circuit of HIV. Nat. Genet. 40:466-470. [DOI] [PubMed] [Google Scholar]
- 34.Weinberger, L. S., and T. Shenk. 2007. An HIV feedback resistor: auto-regulatory circuit deactivator and noise buffer. PLoS Biol. 5:e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu, X., Y. Li, B. Crise, and S. M. Burgess. 2003. Transcription start regions in the human genome are favored targets for MLV integration. Science 300:1749-1751. [DOI] [PubMed] [Google Scholar]
- 36.Yamamichi, N., K. Inada, C. Furukawa, K. Sakurai, T. Tando, A. Ishizaka, T. Haraguchi, T. Mizutani, M. Fujishiro, R. Shimomura, M. Oka, M. Ichinose, Y. Tsutsumi, M. Omata, and H. Iba. 2009. Cdx2 and the Brm-type SWI/SNF complex cooperatively regulate villin expression in gastrointestinal cells. Exp. Cell Res. 315:1779-1789. [DOI] [PubMed] [Google Scholar]
- 37.Yamamichi, N., K. Inada, M. Ichinose, M. Yamamichi-Nishina, T. Mizutani, H. Watanabe, K. Shiogama, M. Fujishiro, T. Okazaki, N. Yahagi, T. Haraguchi, S. Fujita, Y. Tsutsumi, M. Omata, and H. Iba. 2007. Frequent loss of Brm expression in gastric cancer correlates with histologic features and differentiation state. Cancer Res. 67:10727-10735. [DOI] [PubMed] [Google Scholar]
- 38.Yamamichi-Nishina, M., T. Ito, T. Mizutani, N. Yamamichi, H. Watanabe, and H. Iba. 2003. SW13 cells can transition between two distinct subtypes by switching expression of BRG1 and Brm genes at the post-transcriptional level. J. Biol. Chem. 278:7422-7430. [DOI] [PubMed] [Google Scholar]
- 39.Zhang, Z., A. Klatt, D. S. Gilmour, and A. J. Henderson. 2007. Negative elongation factor NELF represses human immunodeficiency virus transcription by pausing the RNA polymerase II complex. J. Biol. Chem. 282:16981-16988. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.