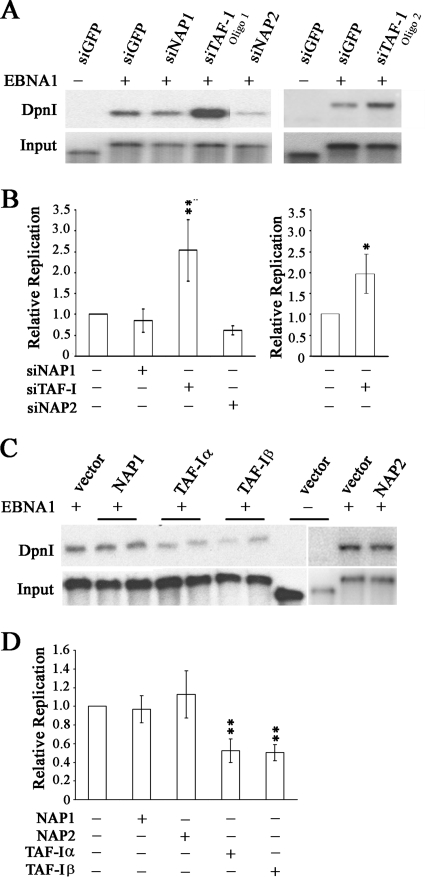
FIG. 5.
Effects of nucleosome assembly proteins on DNA replication activity of EBNA1. (A) CNE2Z cells were treated with siRNA against NAP1, NAP2, TAF-I, or GFP (negative control) and then transfected with an oriP plasmid expressing EBNA1 or lacking EBNA1 (first lane only). For TAF-I, two different siRNA sequences were used (oligo 1 and oligo 2). Three days later, oriP plasmids were harvested from the cells and linearized. One-tenth of the sample was saved as a recovery control (input), and then the remaining sample was digested with DpnI to digest any transfected plasmid that had not replicated in the human cells. Samples were analyzed by Southern blotting and probing for the oriP plasmid. The DpnI-resistant plasmid band is shown in the top panel. (B) Quantification of multiple experiments performed as in panel A. For TAF-I, the left panel is for oligo 1 and the right panel is for oligo 2. Values are shown relative to that for the sample with EBNA1 and siGFP within each experiment, which was set to 1.0. P values of <0.01 (**) and <0.05 (*) are indicated. (C) CNE2Z cells were transfected with plasmids overexpressing NAP1, NAP2, TAF-Iα, or TAF-Iβ or with empty plasmid (vector) along with the oriP plasmid expressing EBNA1 or lacking EBNA1. Replication of the oriP plasmids was then determined as in panel A. (D) Quantification of multiple experiments performed as in panel C. Values are shown relative to that for EBNA1 with an empty overexpression plasmid within each experiment, which was set to 1.0, and P values of <0.01 are indicated.