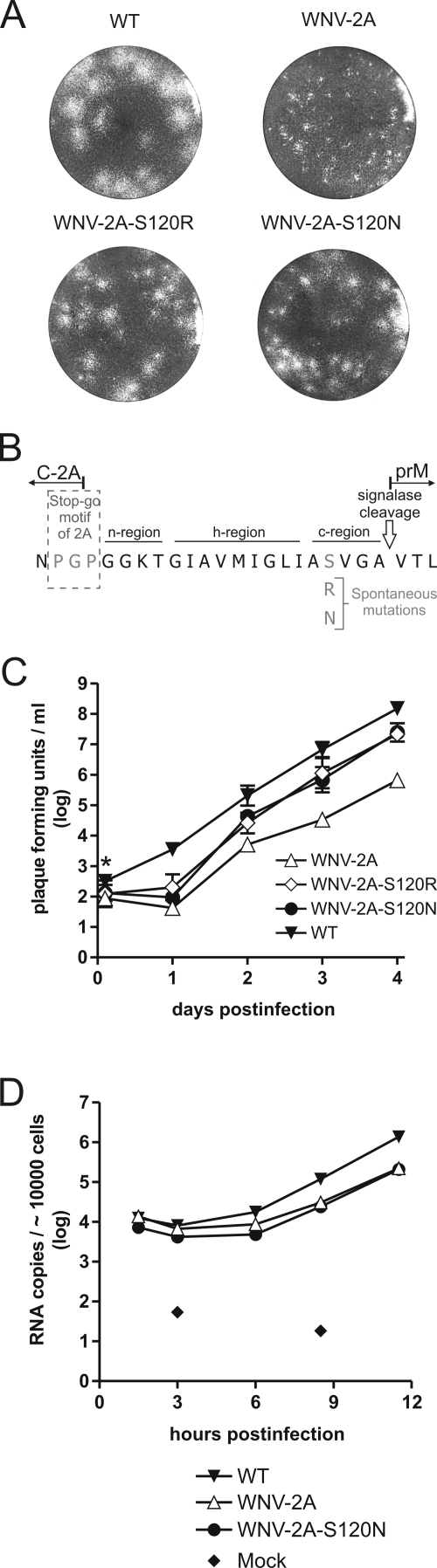
FIG. 8.
Characterization of WNV-2A second-site revertants. (A) Plaque phenotype analysis of WT WNV, WNV-2A, and second-site revertants. Vero cells were infected with cell culture supernatants from the respective viruses. At 4 days postinfection, the cells were stained with crystal violet, and the plaque phenotypes were analyzed. (B) Acquired spontaneous mutations in the signal sequence for prM. The amino acid sequence around the WNV-2A C-prM junction is shown. The n, h, and c regions of the signal sequence for prM are indicated. (C) Growth curve analysis of the indicated viruses in C6/36 cells. The cells were infected at an MOI of 0.01. Samples were taken at different time points, and the infectivity titers were determined using a plaque assay. All data points represent geometric mean values from two independent experiments. The error bars indicate the standard deviations. Inocula that were not completely removed by the washing steps are indicated by an asterisk. (D) RNA replication of WT WNV, WNV-2A, and WNV-2A-S120N after infection of Vero cells. The cells were infected at an MOI of 10. Samples were taken at different time points, and the intracellular RNA levels were measured. All data points represent geometric mean values from two independent experiments.