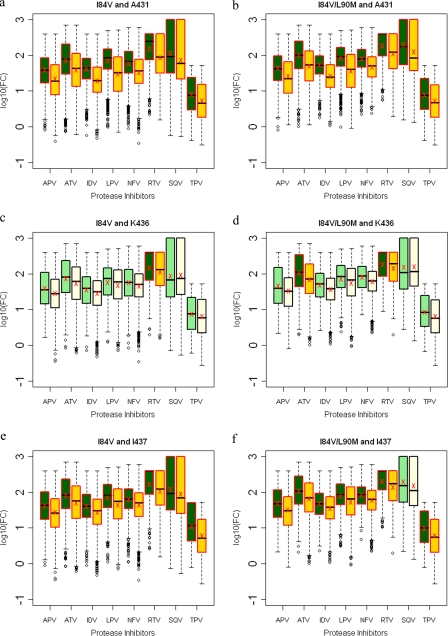
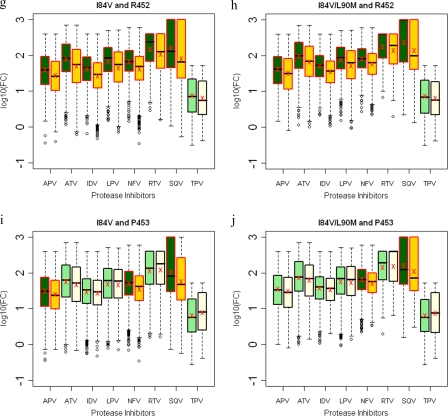
FIG. 9.
Box plots (ratio of median FCs on a log scale; y axis) showing susceptibilities of I84V and I84V/L90M viruses to various PIs (x axis) in the presence (green boxes) or absence (yellow boxes) of associating mutations within the NC-p1 and p1-p6 cleavage sites. The boxes represent the interquartile range between the first and third quartile. The upper and lower lines represent 1.5 times the upper and lower quartile limits. The black line represents the median, and the red X represents the mean of the distribution. Outliers are shown as open circles. Dark green and gold boxes indicate a significant effect of associating mutations within Gag cleavage sites on PI susceptibilities. Susceptibilities are shown as follows: viruses with mutation I84V with and without mutations at Gag 431 (a), viruses with mutation I84V/L90M with and without mutations at Gag 431 (b), viruses with mutation I84V with and without mutations at Gag 436 (c), viruses with mutation I84V/L90M with and without mutations at Gag 436 (d), viruses with mutation I84V with and without mutations at Gag 437 (e), viruses with mutation I84V/L90M with and without mutations at Gag 437 (f), viruses with mutation I84V with and without mutations at Gag 452 (g), viruses with mutation I84V/L90M with and without mutations at 452 (h), viruses with mutation I84V with and without mutations at Gag 453 (i), and viruses with mutation I84V/L90M with and without mutations at Gag 453 (j). P values of ≤0.05 are significant.