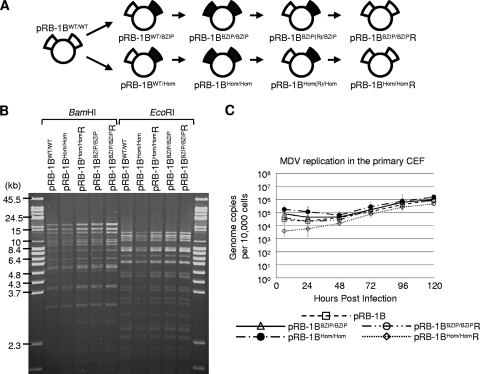
FIG. 5.
Insertion of the mutations into pRB-1B5. (A) Schematic showing the construction of the BAC clones used in the animal experiments: the wild type (pRB-1B5), a double-homodimerization mutant (pRB-1BHom/Hom), and its full revertant (pRB-1BHom/HomR). Also shown are the double-dimerization-null mutant clone (pRB-1BBZIP/BZIP) and its full revertant (pRB-1BBZIP/BZIPR). The intermediate constructs that were not used in the animal experiments are also illustrated (pRB-1BWT/Hom, pRB-1BHom(R)/Hom, pRB-1BWT/BZIP, and pRB-1BBZIP(R)/BZIP). (B) To determine whether the markerless recombination had made any significant second-site mutations or rearrangements, BAC DNA was digested with BamHI or EcoRI restriction enzyme and analyzed by ethidium bromide-stained 1% agarose pulsed-field gel electrophoresis. All of the recombinant MDV BACs were indistinguishable from the wild type. (C) To compare the in vitro replication of the mutant and revertant viruses with that of the parental pRB1B BAC, the genome copy number per 10,000 cells was determined at the indicated times by TaqMan real-time qPCR after inoculation of 100 PFU of each virus into primary CEF. This showed there were no significant differences between the parental and recombinant viruses. The standard error of the mean for each group is shown.