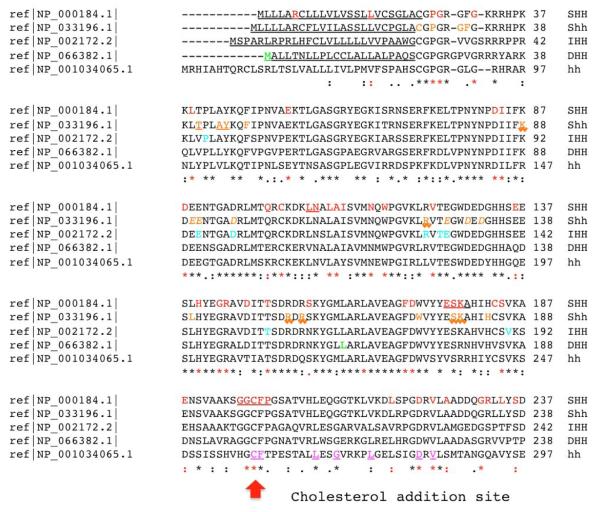
Figure 2. A detailed annotation of known mutations within closely related Hedgehog proteins.
A clustalW2 based (http://www.ebi.ac.uk/Tools/clustalw2/) single aminoacid code alignment of SHH, murine Shh, IHH, DHH and hh is presented. Protein changes are annotated by color and font. For example, HPE or SCMMI alterations are red mutations: normal font are typical missense, red underlined positions are in-frame deleted residues in functionally important motifs, while in-frame deletions or insertions of unknown effect are depicted as black lines above the alignment (p.402_409del and p.405_409del) or as a triangle (i.e. p.Gly411dup). For the mouse Shh alignment we use orange residues with normal font as synthetically engineered point mutations (see Hall et al., 1997; Goetz et al., 2006; Singh et al. 2008), and surface residues implicated in Shh-N-CDOFn3 (ligand-receptor) binding (McLellan et al. 2008) are underlined with a saw-tooth font. In contrast, residues implicated in binding to the fly paralog, Ihog, are orange and underlined. These authors also identified residues that co-ordinate with calcium and promote receptor binding that we present as orange and italic. In addition, IHH (Brachydactyly A-1 or acrofemoralcapital dysplasia; turquoise mutations), DHH (gonadal dysgenesis; green mutations) are presented and compared directly to the Drosophila hh reference sequence. Each paralog encodes a signal peptide shown as underlined normal font (the signal peptide of hh is not shown) and a Sterol Recognition Region (SSR) that is both underlined and italicized. Note that key motifs necessary for hh auto-processing [e.g. the Shh Hint motif (p.258_408) consists of functionally conserved core aminoacids that are show in purple and underlined; a subset of these have been shown to be functionally essential and are double underlined: D303, T326 and H329 (Hall et al. 1997).